-Search query
-Search result
Showing all 35 items for (author: del & campo & r)

EMDB-41785: 
DdmD dimer in complex with ssDNA
Method: single particle / : Bravo JPK, Taylor DW

EMDB-41790: 
DdmD monomer in complex with ssDNA
Method: single particle / : Bravo JPK, Taylor DW

EMDB-41865: 
DdmDE handover complex
Method: single particle / : Bravo JPK, Taylor DW
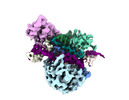
EMDB-41781: 
DdmE in complex with guide and target DNA
Method: single particle / : Bravo JPK, Taylor DW

EMDB-44825: 
DdmD dimer apoprotein (CASP target)
Method: single particle / : Bravo JPK, Taylor DW

EMDB-17231: 
Structure of the FloA-NfeD dimer
Method: single particle / : Ukleja M, Kricks L, Torrens G, Peschiera I, Rodrigues-Lopes I, Krupka M, Garcia Fernandez J, del Campo R, Eulalio A, Mateus A, Lopez Bravo M, Rico AI, Cava F, Lopez D

EMDB-17233: 
Structure of the FloA-NfeD complex with Pbp2a pray bound
Method: single particle / : Ukleja M, Kricks L, Torrens G, Peschiera I, Rodrigues-Lopes I, Krupka M, Garcia Fernandez J, del Campo R, Eulalio A, Mateus A, Lopez Bravo M, Rico AI, Cava F, Lopez D
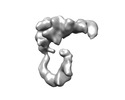
EMDB-17217: 
Structure of Staphylococcus aureus FloA protein
Method: single particle / : Ukleja M, Kricks L, Torrens G, Peschiera I, Rodrigues-Lopes I, Krupka M, Garcia Fernandez J, del Campo R, Eulalio A, Mateus A, Lopez Bravo M, Rico AI, Cava F, Lopez D

EMDB-17222: 
Structure of the FloA-NfeD complex
Method: single particle / : Ukleja M, Kricks L, Torrens G, Peschiera I, Rodrigues-Lopes I, Krupka M, Garcia Fernandez J, del Campo R, Eulalio A, Mateus A, Lopez Bravo M, Rico AI, Cava F, Lopez D

EMDB-17819: 
XBB 1.0 RBD bound to P4J15 (Local)
Method: single particle / : Duhoo Y, Lau K
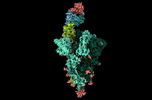
EMDB-17849: 
XBB 1.0 RBD bound to P4J15 (Global)
Method: single particle / : Duhoo Y, Lau K

EMDB-17850: 
SARS-CoV-2 XBB 1.0 closed conformation.
Method: single particle / : Duhoo Y, Lau K
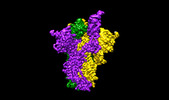
EMDB-17576: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL
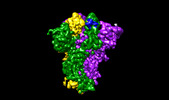
EMDB-17578: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

PDB-8p99: 
SARS-CoV-2 S-protein:D614G mutant in 1-up conformation
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

PDB-8p9y: 
SARS-CoV-2 S protein S:D614G mutant in 3-down with binding site of an entry inhibitor
Method: single particle / : Adhav A, Forcada-Nadal A, Marco-Marin C, Lopez-Redondo ML, Llacer JL

EMDB-14141: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

EMDB-14142: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D
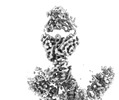
EMDB-14143: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qti: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - 3-P2G3 and 1-P5C3 Fabs (Global)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qtj: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD up - 1-P2G3 and 1-P5C3 Fabs (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

PDB-7qtk: 
SARS-CoV-2 S Omicron Spike B.1.1.529 - RBD down - 1-P2G3 Fab (Local)
Method: single particle / : Ni D, Lau K, Turelli P, Fenwick C, Perez L, Pojer F, Stahlberg H, Pantaleo G, Trono D

EMDB-13190: 
P5C3 is a potent fab neutralizer
Method: single particle / : perez L

EMDB-13265: 
the local resolution of Fab p5c3.
Method: single particle / : perez L

EMDB-13415: 
MaP OF P5C3RBD Interface
Method: single particle / : Perez L

EMDB-0323: 
Structural insights into the ability of nucleoplasmin to assemble and chaperone histone octamers for DNA deposition
Method: single particle / : Valpuesta JM, Arranz R, Martin-Benito J
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search












 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
