-Search query
-Search result
Showing 1 - 50 of 2,298 items for (author: zheng & c)
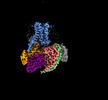
EMDB-60628: 
Carazolol-activated human beta3 adrenergic receptor
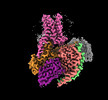
EMDB-60629: 
Epinephrine-activated human beta3 adrenergic receptor

EMDB-60100: 
SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs

EMDB-60101: 
SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, head-to-head aggregate

EMDB-60102: 
SARS-CoV-2 spike trimer (6P) in complex with R1-26 Fab, focused refinement of RBD-Fab region

EMDB-60103: 
SARS-CoV-2 spike trimer (6P) in complex with two H18 Fabs

EMDB-60104: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs

EMDB-60105: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C1 symmetry)

EMDB-60106: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 Fabs, head-to-head aggregate (C3 symmetry)

EMDB-60107: 
SARS-CoV-2 spike trimer (6P) in complex with two H18 and two R1-32 Fabs

EMDB-60108: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs

EMDB-60109: 
SARS-CoV-2 spike trimer (6P) in complex with three H18 and three R1-32 Fabs (one RBD rotated)

EMDB-60110: 
SARS-CoV-2 S1 in complex with H18 and R1-32 Fab

EMDB-60111: 
Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs

EMDB-38845: 
Icosahedrally averaged cryo-EM reconstruction of PhiKZ capsid before applying the "block-based" reconstruction method

EMDB-38846: 
Block 1 of PhiKZ capsid

EMDB-38848: 
Block 2 of PhiKZ capsid

EMDB-39002: 
Composite cryo-EM map of PhiKZ capsid after applying the "block-based" reconstruction method

PDB-8y6v: 
Near-atomic structure of icosahedrally averaged jumbo bacteriophage PhiKZ capsid
EMDB-38080: 
SIRM reconstruction of the MC-45 de novo processed ribosome 50S
EMDB-38081: 
Conventional Reconstruction of the MC-45 de novo processed ribosome 50S
EMDB-38082: 
SIRM reconstruction of the unpublished protein

EMDB-38083: 
The SIRM reconstruction of the MC-40 de novo processed HA-trimer
EMDB-38084: 
The conventional reconstruction of the MC-40 de novo processed HA-trimer

EMDB-38085: 
The SIRM reconstruction of the MC-45 de novo processed PS1

EMDB-38086: 
The conventional reconstruction of the MC-45 de novo processed PS1

EMDB-39299: 
Human resource SGLT1-MAP17 complex
EMDB-38099: 
Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by intein-based E2-Ub-NCP conjugation strategy
EMDB-38100: 
Cryo-EM structures of RNF168/UbcH5c-Ub/nucleosomes complex determined by activity-based chemical trapping strategy
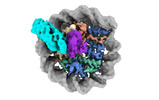
EMDB-38101: 
Cryo-EM structures of RNF168/UbcH5c-Ub in complex with H2AK13Ub nucleosomes determined by activity-based chemical trapping strategy (adjacent H2AK13/15 dual-monoubiquitination)
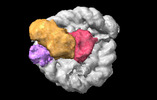
EMDB-38102: 
Cryo-EM map of RNF168/UbcH5c-Ub/nucleosome determined by E2-Ub-NCP conjugation strategy
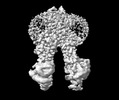
EMDB-38532: 
Cryo-EM structure of human ABCC4

PDB-8xok: 
Cryo-EM structure of human ABCC4

EMDB-37467: 
SARS-CoV-2 Omicron BQ.1.1 RBD complexed with human ACE2

EMDB-37468: 
SARS-CoV-2 Omicron BQ.1 RBD complexed with human ACE2

EMDB-37469: 
SARS-CoV-2 Omicron XBB RBD complexed with human ACE2

EMDB-37470: 
SARS-CoV-2 Omicron BF.7 RBD complexed with human ACE2

EMDB-37471: 
SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304

PDB-8wdy: 
SARS-CoV-2 Omicron BQ.1.1 RBD complexed with human ACE2

PDB-8wdz: 
SARS-CoV-2 Omicron BQ.1 RBD complexed with human ACE2

PDB-8we0: 
SARS-CoV-2 Omicron XBB RBD complexed with human ACE2

PDB-8we1: 
SARS-CoV-2 Omicron BF.7 RBD complexed with human ACE2

PDB-8we4: 
SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304

EMDB-32979: 
Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10)

PDB-7x35: 
Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 8A10 (CVB1-A:8A10)

EMDB-43011: 
Phosphorylated, ATP-bound, E1371Q human cystic fibrosis transmembrane conductance regulator (E1371Q-CFTR)

EMDB-43014: 
Phosphorylated, ATP-bound, inhibitor 172-bound E1371Q human cystic fibrosis transmembrane conductance regulator

EMDB-38533: 
Cryo-EM structure of human ABCC4 with ANP bound in NBD1

EMDB-38534: 
Cryo-EM structure of human ABCC4 in complex with ANP-bound in NBD1 and METHOTREXATE

PDB-8xol: 
Cryo-EM structure of human ABCC4 with ANP bound in NBD1
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
