+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Spike protein / RBD / Antibody / Fab / Viral protein / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion ...symbiont-mediated disruption of host tissue / Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / symbiont entry into host cell / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||||||||
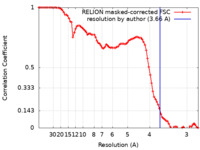
| Method | single particle reconstruction / cryo EM / Resolution: 3.66 Å | ||||||||||||
 Authors Authors | Yan Q / Gao X / Liu B / Hou R / He P / Li Z / Chen Q / Wang J / He J / Chen L ...Yan Q / Gao X / Liu B / Hou R / He P / Li Z / Chen Q / Wang J / He J / Chen L / Zhao J / Xiong X | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants. Authors: Qihong Yan / Xijie Gao / Banghui Liu / Ruitian Hou / Ping He / Yong Ma / Yudi Zhang / Yanjun Zhang / Zimu Li / Qiuluan Chen / Jingjing Wang / Xiaohan Huang / Huan Liang / Huiran Zheng / ...Authors: Qihong Yan / Xijie Gao / Banghui Liu / Ruitian Hou / Ping He / Yong Ma / Yudi Zhang / Yanjun Zhang / Zimu Li / Qiuluan Chen / Jingjing Wang / Xiaohan Huang / Huan Liang / Huiran Zheng / Yichen Yao / Xianying Chen / Xuefeng Niu / Jun He / Ling Chen / Jincun Zhao / Xiaoli Xiong /  Abstract: Continued evolution of SARS-CoV-2 generates variants to challenge antibody immunity established by infection and vaccination. A connection between population immunity and genesis of virus variants ...Continued evolution of SARS-CoV-2 generates variants to challenge antibody immunity established by infection and vaccination. A connection between population immunity and genesis of virus variants has long been suggested but its molecular basis remains poorly understood. Here, we identify a class of SARS-CoV-2 neutralizing public antibodies defined by their shared usage of VL6-57 light chains. Although heavy chains of diverse genotypes are utilized, convergent HCDR3 rearrangements have been observed among these public antibodies to cooperate with germline VL6-57 LCDRs to target a convergent epitope defined by RBD residues S371-S373-S375. Antibody repertoire analysis identifies that this class of VL6-57 antibodies is present in SARS-CoV-2-naive individuals and is clonally expanded in most COVID-19 patients. We confirm that Omicron-specific substitutions at S371, S373 and S375 mediate escape of antibodies of the VL6-57 class. These findings support that this class of public antibodies constitutes a potential immune pressure promoting the introduction of S371L/F-S373P-S375F in Omicron variants. The results provide further molecular evidence to support that antigenic evolution of SARS-CoV-2 is driven by antibody mediated population immunity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60111.map.gz emd_60111.map.gz | 48.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60111-v30.xml emd-60111-v30.xml emd-60111.xml emd-60111.xml | 27.6 KB 27.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60111_fsc.xml emd_60111_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_60111.png emd_60111.png | 84.7 KB | ||
| Filedesc metadata |  emd-60111.cif.gz emd-60111.cif.gz | 7.8 KB | ||
| Others |  emd_60111_half_map_1.map.gz emd_60111_half_map_1.map.gz emd_60111_half_map_2.map.gz emd_60111_half_map_2.map.gz | 48.8 MB 48.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60111 http://ftp.pdbj.org/pub/emdb/structures/EMD-60111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60111 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60111 | HTTPS FTP |
-Validation report
| Summary document |  emd_60111_validation.pdf.gz emd_60111_validation.pdf.gz | 829.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60111_full_validation.pdf.gz emd_60111_full_validation.pdf.gz | 829.4 KB | Display | |
| Data in XML |  emd_60111_validation.xml.gz emd_60111_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_60111_validation.cif.gz emd_60111_validation.cif.gz | 19.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60111 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60111 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60111 | HTTPS FTP |
-Related structure data
| Related structure data |  8zhpMC  8zhdC  8zheC  8zhfC  8zhgC  8zhhC  8zhiC  8zhjC  8zhkC  8zhlC  8zhmC  8zhnC  8zhoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60111.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60111.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.395 Å | ||||||||||||||||||||||||||||||||||||
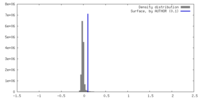
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_60111_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
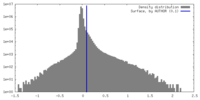
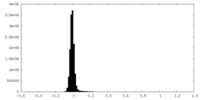
| Density Histograms |
-Half map: #1
| File | emd_60111_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
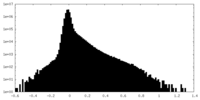
| Density Histograms |
- Sample components
Sample components
+Entire : Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs
+Supramolecule #1: Dimer of SARS-CoV-2 S1 in complex with H18 and R1-32 Fabs
+Supramolecule #2: SARS-CoV-2 S1
+Supramolecule #3: H18 Fab
+Supramolecule #4: R1-32 Fab
+Macromolecule #1: Spike glycoprotein
+Macromolecule #2: Heavy chain of R1-32 Fab
+Macromolecule #3: Light chain of R1-32 Fab
+Macromolecule #4: Light chain of H18 Fab
+Macromolecule #5: Heavy chain of H18 Fab
+Macromolecule #6: 2-acetamido-2-deoxy-beta-D-glucopyranose
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)