+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human ABCC4 with ANP bound in NBD1 | |||||||||
 Map data Map data | C4AMP | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ABC transporter MRP / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information15-hydroxyprostaglandin dehydrogenase (NAD+) activity / purine nucleotide transmembrane transporter activity / cAMP transport / guanine nucleotide transmembrane transporter activity / ABC-type bile acid transporter activity / platelet dense granule membrane / platelet degranulation / leukotriene transport / urate transport / ABC-type glutathione S-conjugate transporter activity ...15-hydroxyprostaglandin dehydrogenase (NAD+) activity / purine nucleotide transmembrane transporter activity / cAMP transport / guanine nucleotide transmembrane transporter activity / ABC-type bile acid transporter activity / platelet dense granule membrane / platelet degranulation / leukotriene transport / urate transport / ABC-type glutathione S-conjugate transporter activity / prostaglandin transport / ABC-type glutathione-S-conjugate transporter / glutathione transmembrane transporter activity / prostaglandin transmembrane transporter activity / : / urate transmembrane transporter activity / external side of apical plasma membrane / xenobiotic transmembrane transport / prostaglandin secretion / Paracetamol ADME / export across plasma membrane / ABC-type xenobiotic transporter / Translocases; Catalysing the translocation of other compounds; Linked to the hydrolysis of a nucleoside triphosphate / Azathioprine ADME / ABC-type xenobiotic transporter activity / bile acid and bile salt transport / efflux transmembrane transporter activity / cilium assembly / ATPase-coupled transmembrane transporter activity / xenobiotic transmembrane transporter activity / ABC-type transporter activity / transport across blood-brain barrier / xenobiotic metabolic process / ABC-family proteins mediated transport / transmembrane transport / Platelet degranulation / basolateral plasma membrane / apical plasma membrane / intracellular membrane-bounded organelle / nucleolus / Golgi apparatus / ATP hydrolysis activity / ATP binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.02 Å | |||||||||
 Authors Authors | Zhang PF / Liu Z | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: FEBS Lett / Year: 2024 Journal: FEBS Lett / Year: 2024Title: The ATP-bound inward-open conformation of ABCC4 reveals asymmetric ATP binding for substrate transport. Authors: Yue Zhu / Xiaoke Xing / Fuxing Wang / Luojun Chen / Chunhui Zhong / Xiting Lu / Zhanwang Yu / Yongbo Yang / Yi Yao / Qibin Song / Suxia Han / Zheng Liu / Pingfeng Zhang /  Abstract: The multidrug resistance-associated protein (MRP) ABCC4 facilitates substrate transport across the cytoplasmic membrane, crucial for normal physiology and mediating multidrug resistance in tumor ...The multidrug resistance-associated protein (MRP) ABCC4 facilitates substrate transport across the cytoplasmic membrane, crucial for normal physiology and mediating multidrug resistance in tumor cells. Despite intensive studies on MRPs, ABCC4's transport mechanism remains incompletely understood. In this study, we unveiled an inward-open conformation with an ATP bound to degenerate NBD1. Additionally, we captured the structure with both ATP and substrate co-bound in the inward-open state. Our findings uncover the asymmetric ATP binding in ABCC4 and provide insights into substrate binding and transport mechanisms. ATP binding to NBD1 is parallel to substrate binding to ABCC4, and is a prerequisite for ATP-bound NBD2-induced global conformational changes. Our findings shed new light on targeting ABCC4 in combination with anticancer therapy. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38533.map.gz emd_38533.map.gz | 31.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38533-v30.xml emd-38533-v30.xml emd-38533.xml emd-38533.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38533.png emd_38533.png | 78.4 KB | ||
| Filedesc metadata |  emd-38533.cif.gz emd-38533.cif.gz | 6.9 KB | ||
| Others |  emd_38533_half_map_1.map.gz emd_38533_half_map_1.map.gz emd_38533_half_map_2.map.gz emd_38533_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38533 http://ftp.pdbj.org/pub/emdb/structures/EMD-38533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38533 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38533 | HTTPS FTP |
-Related structure data
| Related structure data |  8xolMC  8xokC  8xomC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38533.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38533.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C4AMP | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
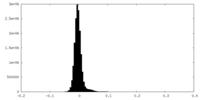
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: C4AMP half map B
| File | emd_38533_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C4AMP half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
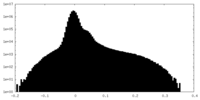
| Density Histograms |
-Half map: C4AMP half map A
| File | emd_38533_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | C4AMP half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
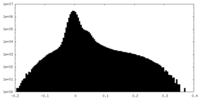
| Density Histograms |
- Sample components
Sample components
-Entire : human ABCC4 in complex with ANP-bound in NBD1
| Entire | Name: human ABCC4 in complex with ANP-bound in NBD1 |
|---|---|
| Components |
|
-Supramolecule #1: human ABCC4 in complex with ANP-bound in NBD1
| Supramolecule | Name: human ABCC4 in complex with ANP-bound in NBD1 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 149.5 KDa |
-Macromolecule #1: ATP-binding cassette sub-family C member 4
| Macromolecule | Name: ATP-binding cassette sub-family C member 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: ABC-type xenobiotic transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 149.693922 KDa |
| Recombinant expression | Organism: Mammalian expression vector Flag-MCS-pcDNA3.1 (others) |
| Sequence | String: MLPVYQEVKP NPLQDANLCS RVFFWWLNPL FKIGHKRRLE EDDMYSVLPE DRSQHLGEEL QGFWDKEVLR AENDAQKPSL TRAIIKCYW KSYLVLGIFT LIEESAKVIQ PIFLGKIINY FENYDPMDSV ALNTAYAYAT VLTFCTLILA ILHHLYFYHV Q CAGMRLRV ...String: MLPVYQEVKP NPLQDANLCS RVFFWWLNPL FKIGHKRRLE EDDMYSVLPE DRSQHLGEEL QGFWDKEVLR AENDAQKPSL TRAIIKCYW KSYLVLGIFT LIEESAKVIQ PIFLGKIINY FENYDPMDSV ALNTAYAYAT VLTFCTLILA ILHHLYFYHV Q CAGMRLRV AMCHMIYRKA LRLSNMAMGK TTTGQIVNLL SNDVNKFDQV TVFLHFLWAG PLQAIAVTAL LWMEIGISCL AG MAVLIIL LPLQSCFGKL FSSLRSKTAT FTDARIRTMN EVITGIRIIK MYAWEKSFSN LITNLRKKEI SKILRSSCLR GMN LASFFS ASKIIVFVTF TTYVLLGSVI TASRVFVAVT LYGAVRLTVT LFFPSAIERV SEAIVSIRRI QTFLLLDEIS QRNR QLPSD GKKMVHVQDF TAFWDKASET PTLQGLSFTV RPGELLAVVG PVGAGKSSLL SAVLGELAPS HGLVSVHGRI AYVSQ QPWV FSGTLRSNIL FGKKYEKERY EKVIKACALK KDLQLLEDGD LTVIGDRGTT LSGGQKARVN LARAVYQDAD IYLLDD PLS AVDAEVSRHL FELCICQILH EKITILVTHQ LQYLKAASQI LILKDGKMVQ KGTYTEFLKS GIDFGSLLKK DNEESEQ PP VPGTPTLRNR TFSESSVWSQ QSSRPSLKDG ALESQDTENV PVTLSEENRS EGKVGFQAYK NYFRAGAHWI VFIFLILL N TAAQVAYVLQ DWWLSYWANK QSMLNVTVNG GGNVTEKLDL NWYLGIYSGL TVATVLFGIA RSLLVFYVLV NSSQTLHNK MFESILKAPV LFFDRNPIGR ILNRFSKDIG HLDDLLPLTF LDFIQTLLQV VGVVSVAVAV IPWIAIPLVP LGIIFIFLRR YFLETSRDV KRLESTTRSP VFSHLSSSLQ GLWTIRAYKA EERCQELFDA HQDLHSEAWF LFLTTSRWFA VRLDAICAMF V IIVAFGSL ILAKTLDAGQ VGLALSYALT LMGMFQWCVR QSAEVENMMI SVERVIEYTD LEKEAPWEYQ KRPPPAWPHE GV IIFDNVN FMYSPGGPLV LKHLTALIKS QEKVGIVGRT GAGKSSLISA LFRLSEPEGK IWIDKILTTE IGLHDLRKKM SII PQEPVL FTGTMRKNLD PFNEHTDEEL WNALQEVQLK ETIEDLPGKM DTELAESGSN FSVGQRQLVC LARAILRKNQ ILII DEATA NVDPRTDELI QKKIREKFAH CTVLTIAHRL NTIIDSDKIM VLDSGRLKEY DEPYVLLQNK ESLFYKMVQQ LGKAE AAAL TETAKQVYFK RNYPHIGHTD HMVTNTSNGQ PSTLTIFETA L UniProtKB: ATP-binding cassette sub-family C member 4 |
-Macromolecule #2: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 2 / Number of copies: 1 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #3: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13...
| Macromolecule | Name: 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol type: ligand / ID: 3 / Number of copies: 8 / Formula: DU0 |
|---|---|
| Molecular weight | Theoretical: 516.752 Da |
| Chemical component information |  ChemComp-DU0: |
-Macromolecule #4: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 4 / Number of copies: 1 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Component - Concentration: 25.0 mM / Component - Name: Hepes Details: 50 mM HEPES pH 7.5, 150 mM NaCl, 2 mM DTT, 2 mM MgCl2, 0.005% (w/v) GDN and 0.0005% CHS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: After incubation on the grids at 277K under 100% humidity for 10 s, the grids were bloted for 3.0 s and then plunged frozen into liquid ethane cooled by liquid nitrogen using a Vitrobot.. |
| Details | The human ABCC4 protein was purified in 0.005% (w/v) GDN and 0.0005% CHS, the peak protein fraction from gel filtration column were concentrated to about 10 mg/ml. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 55.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8xol: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)