-Search query
-Search result
Showing 1 - 50 of 326 items for (author: xinzheng & z)

EMDB-33625: 
Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

EMDB-33626: 
Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex
Method: single particle / : Liu CP, Yu C, Yu ZY, Xu RM

EMDB-33627: 
Cryo-EM structure of the left-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

EMDB-33630: 
Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu C, Yu ZY, Xu RM
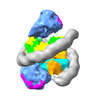
EMDB-33631: 
Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

EMDB-35660: 
Composite cryo-EM density map of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM
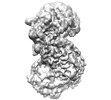
EMDB-35661: 
Cryo-EM structure of the monomeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-35708: 
Cryo-EM consensus map of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-35709: 
Local refinement cryo-EM map of protomer I of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-35710: 
Local refinement cryo-EM map of protomer II of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-36013: 
Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-36014: 
Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Xu RM

PDB-7y5u: 
Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

PDB-7y5v: 
Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex
Method: single particle / : Liu CP, Yu C, Yu ZY, Xu RM

PDB-7y5w: 
Cryo-EM structure of the left-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

PDB-7y60: 
Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu C, Yu ZY, Xu RM

PDB-7y61: 
Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Yu C, Xu RM

PDB-8iqf: 
Cryo-EM structure of the dimeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

PDB-8iqg: 
Cryo-EM structure of the monomeric human CAF1-H3-H4 complex
Method: single particle / : Liu CP, Yu ZY, Xu RM

PDB-8j6s: 
Cryo-EM structure of the single CAF-1 bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Xu RM

PDB-8j6t: 
Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome
Method: single particle / : Liu CP, Yu ZY, Xu RM

EMDB-33027: 
SFTSV 3 fold hexmer
Method: single particle / : Sun Z, Lou Z

EMDB-33030: 
SFTSV 5 fold pentamer
Method: single particle / : Sun Z, Lou Z

EMDB-36343: 
The cryo-EM structure of Parvovirus milled by 30 keV gallium FIB at 3.09 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36346: 
The cryo-EM structure of 10-20nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 4.05 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36347: 
The cryo-EM structure of 20-30nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 3.88 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36348: 
The cryo-EM structure of 30-40 nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 3.82 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36349: 
The cryo-EM structure of 40-50 nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 3.62 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36350: 
The cryo-EM structure of 50-60 nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 3.60 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36351: 
The cryo-EM structure of 60-70 nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 3.57 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ
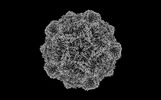
EMDB-36352: 
The cryo-EM structure of Parvovirus milled by 8 keV gallium FIB at 3.11 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36355: 
The cryo-EM structure of 10-20 nm group Parvovirus from lamellae surface milled by 8 keV Ga+ FIB at 3.88 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36356: 
The cryo-EM structure of 20-30 nm group Parvovirus from lamellae surface milled by 8 keV Ga+ FIB at 3.70 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36357: 
The cryo-EM structure of 30-40 nm group Parvovirus from lamellae surface milled by 8 keV Ga+ FIB at 3.60 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36358: 
The cryo-EM structure of 40-50 nm group Parvovirus from lamellae surface milled by 8 keV Ga+ FIB at 3.54 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-36359: 
The cryo-EM structure of 50-60 nm group Parvovirus from lamellae surface milled by 8 keV Ga+ FIB at 3.57 Angstrom resolution.
Method: single particle / : Yang Q, Zhang XZ

EMDB-33029: 
SFTSV 2 fold hexamer
Method: single particle / : Sun Z, Lou Z

EMDB-33669: 
In situ double-PBS-PSII-PSI-LHCs megacomplex from red alga Porphyridium purpureum.
Method: single particle / : You X, Zhang X, Cheng J, Xiao YN, Sun S, Sui SF

EMDB-35976: 
Structure of in situ PSII from P. purpureum lamellae by GisSPA
Method: single particle / : Zhang X, Cheng J

EMDB-34227: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 1
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34229: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 4
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34230: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 2
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34235: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 3
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34236: 
Cryo-EM structure of Singapore Grouper Iridovirus capsid block 5
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF
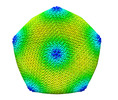
EMDB-34251: 
Singapore Grouper Iridovirus
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

EMDB-34815: 
One asymmetric unit of Singapore grouper iridovirus capsid
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF

PDB-8hif: 
One asymmetric unit of Singapore grouper iridovirus capsid
Method: single particle / : Zhao ZN, Liu CC, Zhu DJ, Qi JX, Zhang XZ, Gao GF
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search






 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
