[English] 日本語
 Yorodumi
Yorodumi- EMDB-36348: The cryo-EM structure of 30-40 nm group Parvovirus from lamellae ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of 30-40 nm group Parvovirus from lamellae surface milled by 30 keV Ga+ FIB at 3.82 Angstrom resolution. | |||||||||
 Map data Map data | CryoEM structure of 30-40 nm group parvovirus from lamellae surface milled by 30 keV Ga FIB. Block-based reconstruction. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | icosahedral / VIRUS LIKE PARTICLE | |||||||||
| Biological species |  Protoparvovirus Protoparvovirus | |||||||||
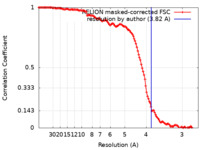
| Method | single particle reconstruction / cryo EM / Resolution: 3.82 Å | |||||||||
 Authors Authors | Yang Q / Zhang XZ | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2023 Journal: Structure / Year: 2023Title: The reduction of FIB damage on cryo-lamella by lowering energy of ion beam revealed by a quantitative analysis. Authors: Qi Yang / Chunling Wu / Dongjie Zhu / Junxi Li / Jing Cheng / Xinzheng Zhang /  Abstract: Focused ion beam (FIB) is widely used for thinning frozen cells to produce lamellae for cryo-electron microscopy imaging and for protein structures study in vivo. However, FIB damages the lamellae ...Focused ion beam (FIB) is widely used for thinning frozen cells to produce lamellae for cryo-electron microscopy imaging and for protein structures study in vivo. However, FIB damages the lamellae and a quantitative experimental analysis of the damage is lacking. We used a 30-keV gallium FIB to prepare lamellae of a highly concentrated icosahedral virus sample. The viruses were grouped according to their distance from the surface of lamellae and reconstructed. Damage to the approximately 20-nm-thick outermost lamella surface was similar to that from exposure to 16 e/Å in a 300-kV cryo-electron microscope at high-resolution range. The damage was negligible at a depth beyond 50 nm, which was reduced to 30 nm if 8-keV Ga was used during polishing. We designed extra steps in the reconstruction refinement to maximize undamaged signals and increase the resolution. The results demonstrated that low-energy beam polishing was essential for high-quality thinner lamellae. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36348.map.gz emd_36348.map.gz | 164.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36348-v30.xml emd-36348-v30.xml emd-36348.xml emd-36348.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36348_fsc.xml emd_36348_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_36348.png emd_36348.png | 130.8 KB | ||
| Filedesc metadata |  emd-36348.cif.gz emd-36348.cif.gz | 3.9 KB | ||
| Others |  emd_36348_half_map_1.map.gz emd_36348_half_map_1.map.gz emd_36348_half_map_2.map.gz emd_36348_half_map_2.map.gz | 141.2 MB 141.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36348 http://ftp.pdbj.org/pub/emdb/structures/EMD-36348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36348 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36348 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36348.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36348.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM structure of 30-40 nm group parvovirus from lamellae surface milled by 30 keV Ga FIB. Block-based reconstruction. | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 2 of 30-40 nm group parvovirus...
| File | emd_36348_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of 30-40 nm group parvovirus from lamellae surface milled by 30 keV Ga FIB. Block-based reconstruction. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1 of 30-40 nm group parvovirus...
| File | emd_36348_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of 30-40 nm group parvovirus from lamellae surface milled by 30 keV Ga FIB. Block-based reconstruction. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Protoparvovirus
| Entire | Name:  Protoparvovirus Protoparvovirus |
|---|---|
| Components |
|
-Supramolecule #1: Protoparvovirus
| Supramolecule | Name: Protoparvovirus / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 1506574 / Sci species name: Protoparvovirus / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: OTHER / Virus enveloped: No / Virus empty: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 70 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 2.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)