-Search query
-Search result
Showing 1 - 50 of 172 items for (author: wang & dn)

EMDB-53311: 
Cryo-EM map of SKM-70S ribosomal stalled complex in the major state (vacant A-site, canon)
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

EMDB-53341: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the A-tRNA positioned (Body open) state.
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

EMDB-55145: 
Cryo-EM structure of SKM-70S ribosomal stalled complex in the rotated state with hybrid tRNAs
Method: single particle / : Morici M, Corazza M, Safdari HA, Wilson DN

EMDB-39244: 
Rhodobacter blasticus RC-LH1 dimer
Method: single particle / : Liu LN, Zhang YZ, Wang P, Christianson BM, Ugurlar D

EMDB-39255: 
Rhodobacter blasticus RC-LH1 monomer
Method: single particle / : Liu LN, Zhang YZ, Wang P, Christianson BM, Ugurlar D

PDB-8ygd: 
Rhodobacter blasticus RC-LH1 dimer
Method: single particle / : Liu LN, Zhang YZ, Wang P, Christianson BM, Ugurlar D

PDB-8ygl: 
Rhodobacter blasticus RC-LH1 monomer
Method: single particle / : Liu LN, Zhang YZ, Wang P, Christianson BM, Ugurlar D

EMDB-43571: 
Structure of the five-fold capsomer of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

EMDB-43573: 
Structure of the three-fold capsomer of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

EMDB-43575: 
Structure of the non-symmetric capsomer of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

EMDB-43584: 
Structure of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

PDB-8vvw: 
Structure of the five-fold capsomer of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

PDB-8vvz: 
Structure of the three-fold capsomer of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

PDB-8vw3: 
Structure of the non-symmetric capsomer of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA

PDB-8vwg: 
Structure of the Drosophila retrotransposon Copia capsid
Method: single particle / : Liu Y, Kelch BA
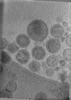
EMDB-47705: 
Tomogram of Cas9 containing Enveloped Delivery Vehicles (EDVs)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J
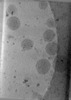
EMDB-47741: 
Tomogram of Enveloped Delivery Vehicles (EDVs) without Cas9
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47743: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) without Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-47745: 
Subtomogram average of capsid (CA) from immature Enveloped Delivery Vehicles (EDVs) containing Cas9
Method: subtomogram averaging / : Peukes J, Ngo W, Nogales E, Doudna J
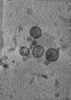
EMDB-47858: 
Tomogram of a minimized Enveloped Delivery Vehicle (miniEDV)
Method: electron tomography / : Peukes J, Ngo W, Nogales E, Doudna J

EMDB-42614: 
Structure of NaCT-PF4a complex
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42615: 
Structure of NaDC3-aKG complex
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42616: 
Structure of NaDC3-PF4a complex
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42617: 
Structure of NaDC3-Succinate complex in Coo-Ci conformation
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42618: 
Structure of NaDC3-DMS complex in Ci-Ci conformation
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42619: 
Structure of NaDC3-DMS complex in Co-Ci conformation
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42620: 
Structure of NaCT-succ complex
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-42621: 
Structure of NaDC3-Succinate complex in Coo-Coo conformation
Method: single particle / : Li Y, Wang DN, Mindell JA, Rice WJ, Song J, Mikusevic V, Marden JJ, Becerril A, Kuang H, Wang B

EMDB-44143: 
NorA in complex with Fab36 (NorA-BRIL fusion)
Method: single particle / : Xie P, Li Y, Kuang H, Wang DN, Traaseth NJ

EMDB-44144: 
NorA in occluded conformation (NorA-BRIL fusion)
Method: single particle / : Xie P, Li Y, Kuang H, Wang DN, Traaseth NJ

EMDB-44145: 
NorA in inward-open conformation (NorA-BRIL fusion)
Method: single particle / : Xie P, Li Y, Kuang H, Wang DN, Traaseth NJ

EMDB-44147: 
NorA in inward-occluded conformation (NorA-BRIL fusion)
Method: single particle / : Xie P, Li Y, Kuang H, Wang DN, Traaseth NJ

EMDB-41605: 
Protonated state of NorA at pH 5.0
Method: single particle / : Li JP, Li Y, Koide A, Kuang HH, Torres VJ, Koide S, Wang DN, Traaseth NJ

EMDB-41606: 
NorA double mutant - E222QD307N at pH 7.5
Method: single particle / : Li JP, Li Y, Koide A, Kuang HH, Torres VJ, Koide S, Wang DN, Traaseth NJ

EMDB-41607: 
NorA single mutant - E222Q at pH 7.5
Method: single particle / : Li JP, Li Y, Koide A, Kuang HH, Torres VJ, Koide S, Wang DN, Traaseth NJ

EMDB-41608: 
NorA single mutant - D307N at pH 7.5
Method: single particle / : Li JP, Li Y, Koide A, Kuang HH, Torres VJ, Koide S, Wang DN, Traaseth NJ

EMDB-38453: 
Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

EMDB-38454: 
Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

PDB-8xlm: 
Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein in complex with ACE2 (1-up state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

PDB-8xln: 
Structure of the SARS-CoV-2 EG.5.1 spike RBD in complex with ACE2
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

EMDB-37648: 
SARS-CoV-2 EG.5.1 spike glycoprotein (1-up state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

EMDB-37650: 
SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

EMDB-37651: 
SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

PDB-8wmd: 
Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

PDB-8wmf: 
Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state)
Method: single particle / : Nomai T, Anraku Y, Kita S, Hashiguchi T, Maenaka K

EMDB-41374: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS

EMDB-41382: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS

EMDB-41399: 
Antibody N3-1 bound to SARS-CoV-2 spike
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tm1: 
Antibody N3-1 bound to RBDs in the up and down conformations
Method: single particle / : Hsieh CL, McLellan JS

PDB-8tma: 
Antibody N3-1 bound to RBD in the up conformation
Method: single particle / : Hsieh CL, McLellan JS
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
