-Search query
-Search result
Showing all 48 items for (author: shan & yy)

EMDB-36800: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-36801: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state, focused refined on KtrA octamer
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-36802: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state, focused refined on KtrB dimer
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-36803: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-36804: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-38477: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-38478: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

PDB-8k1s: 
Potassium transporter KtrAB from Bacillus subtilis in ADP-bound state
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

PDB-8k1t: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of MgCl2
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

PDB-8k1u: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

PDB-8xmh: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, vertical C2 symmetry axis
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

PDB-8xmi: 
Potassium transporter KtrAB from Bacillus subtilis in ATP-bound state with addition of EDTA and EGTA, C1 symmetry
Method: single particle / : Chang YK, Chiang WT, Hu NJ, Tsai MD

EMDB-34259: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv282
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34260: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv282 focused on RBD_XGv282 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q

EMDB-34261: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv289
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34262: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv289 focused on RBD_XGv289 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q

EMDB-34263: 
cryo-EM structure of Omicron BA.5 S protein in complex with S2L20
Method: single particle / : Xia XY, Zhang YY, Chi XM, Huang BD, Wu LS, Zhou Q

EMDB-34264: 
Cryo-EM map of Omicron BA.5 S protein in complex with S2L20 focused on NTD_S2L20 sub-complex
Method: single particle / : Xia LY, Zhang YY, Zhou Q

EMDB-33522: 
The pre-fusion structure of Thogotovirus dhori envelope glycoprotein
Method: single particle / : Shan YY, Zhang MF, Pei DQ

EMDB-33451: 
Cryo-EM structure of SLC19A1
Method: single particle / : Zhang MF, Shan YY, Pei DQ

EMDB-32869: 
S protein of Delta variant in complex with ZWD12
Method: single particle / : Guo YY, Zhang YY, Zhou Q

EMDB-32870: 
S protein of Delta variant in complex with ZWD12 focused on RBD_ZWD12 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-32871: 
S protein of Delta variant in complex with ZWC6
Method: single particle / : Guo YY, Zhang YY, Zhou Q

EMDB-32872: 
S protein of Delta variant in complex with ZWC6 focused on RBD_ZWC6 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-32920: 
S protein of SARS-CoV-2 in complex with 2G1
Method: single particle / : Guo YY, Zhang YY

EMDB-32921: 
S protein of SARS-CoV-2 in complex with 2G1 focused on RBD_2G1 sub-complex
Method: single particle / : Guo YY, Zhang YY

EMDB-31146: 
Coupling of N7-methyltransferase and 3'-5' exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading
Method: single particle / : Yan L, Yang YX

EMDB-31138: 
Co-transcriptional capping machineries in SARS-CoV-2 RTC: Coupling of N7-methyltransferase and 3'-5' exoribonuclease with polymerase reveals mechanisms for capping and proofreading
Method: single particle / : Yan LM, Yang YX

EMDB-11942: 
"Tubulin glycylation controls axonemal dynein activity, flagellar beat and male fertility": Pre-pre-power stroke conformation of the mouse sperm outer dynein arms (ODAs). Both beta- and gamma-heavy chains are in pre-power stroke conformation. Cryo-ET, classification and sub-tomogram averaging.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11943: 
"Tubulin glycylation controls axonemal dynein activity, flagellar beat and male fertility": Pre-post-power stroke conformation of the mouse sperm outer dynein arms (ODAs), with gamma-heavy chain in pre-power stroke conformation and beta-heavy chain in post-power stroke conformation. Cryo-ET, classification and sub-tomogram averaging.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11944: 
Post-pre-power stroke conformation of the mouse sperm outer dynein arms (ODAs), with gamma-heavy chain in post-power stroke conformation and beta-heavy chain in pre-power stroke conformation. Reconstruction obtained by sub-tomogram classification and averaging from cryo-electron tomograms of active flagella of mouse sperm cells.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11945: 
Post-post-power stroke conformation of the mouse sperm outer dynein arms (ODAs), with both gamma-heavy chain and beta-heavy chain in post-power stroke conformation. Reconstruction obtained with sub-tomogram classification and averaging from cryo-electron tomograms of active flagella of mouse sperm cells.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11946: 
Axonemal 96nm-repeat from active sperm flagella of wild type mouse. Reconstruction obtained with cryo-ET and sub-tomogram averaging.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11947: 
Axonemal 96nm-repeat from active sperm flagella of glycylation deficient mouse (Ttll3-/-Ttll8-/- mutation). Reconstruction obtained with cryo-ET and sub-tomogram averaging.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11948: 
Axonemal 96nm-repeat from active flagella of wild type mouse, showing inner dynein arms (IDAs) in pre-power stroke conformations. Reconstruction obtained with classification and averaging of sub-tomograms from cryo-electron tomograms of wild type active flagella.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-11949: 
Axonemal 96nm-repeat from active flagella of wild type mouse, showing inner dynein arms (IDAs) in post-power stroke conformations. Reconstruction obtained with classification and averaging of sub-tomograms from cryo-electron tomograms of wild type active flagella.
Method: subtomogram averaging / : Gadadhar S, Alvarez Viar G, Hansen JN, Gong A, Kostarev A, Ialy-Radio C, Leboucher S, Whitfield M, Ziyyat A, Toure A, Alvarez L, Pigino G, Janke C

EMDB-22907: 
SARS-CoV-2 Spike bound to Nb6 in closed conformation
Method: single particle / : Schoof MS, Faust BF

EMDB-22908: 
SARS-CoV-2 Spike bound to Nb6 in open conformation
Method: single particle / : Schoof MS, Faust BF, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesboelle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Desphande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Krogan NJ, Ralston CY, Swaney DL, Garcia-Sastre A, Ott M, Vignuzzi M, Walter P, Manglik A, QCRG Structural Biology Consortium

EMDB-22909: 
SARS-CoV-2 Spike bound to Nb11 in closed conformation
Method: single particle / : Schoof MS, Faust BF, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesboelle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Desphande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Krogan NJ, Ralston CY, Swaney DL, Garcia-Sastre A, Ott M, Vignuzzi M, Walter P, Manglik A, QCRG Structural Biology Consortium

EMDB-22910: 
SARS-CoV-2 Spike bound to mNb6 in closed conformation
Method: single particle / : Schoof MS, Faust BF

EMDB-22911: 
SARS-CoV-2 Spike bound to Nb11 in open conformation
Method: single particle / : Schoof MS, Faust BF, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesboelle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Desphande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Krogan NJ, Ralston CY, Swaney DL, Garcia-Sastre A, Ott M, Vignuzzi M, Walter P, Manglik A, QCRG Structural Biology Consortium

PDB-7kkk: 
SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6
Method: single particle / : Schoof MS, Faust BF, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesboelle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Desphande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, Garcia-Sastre A, Ott M, Vignuzzi M, Walter P, Manglik A, QCRG Structural Biology Consortium

PDB-7kkl: 
SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6
Method: single particle / : Schoof MS, Faust BF, Saunders RA, Sangwan S, Rezelj V, Hoppe N, Boone M, Billesboelle CB, Puchades C, Azumaya CM, Kratochvil HT, Zimanyi M, Desphande I, Liang J, Dickinson S, Nguyen HC, Chio CM, Merz GE, Thompson MC, Diwanji D, Schaefer K, Anand AA, Dobzinski N, Zha BS, Simoneau CR, Leon K, White KM, Chio US, Gupta M, Jin M, Li F, Liu Y, Zhang K, Bulkley D, Sun M, Smith AM, Rizo AN, Moss F, Brilot AF, Pourmal S, Trenker R, Pospiech T, Gupta S, Barsi-Rhyne B, Belyy V, Barile-Hill AW, Nock S, Liu Y, Krogan NJ, Ralston CY, Swaney DL, Garcia-Sastre A, Ott M, Vignuzzi M, Walter P, Manglik A, QCRG Structural Biology Consortium

EMDB-30276: 
cryo EM map of the S protein of SARS-CoV-2 in complex bound with 4A8
Method: single particle / : Yan RH, Zhang YY

EMDB-30277: 
Cryo EM map of the interface between NTD of SARS-CoV-2 and 4A8
Method: single particle / : Yan RH, Zhang YY, Guo YY, Li YN, Xia L, Zhou Q
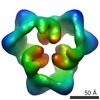
EMDB-1897: 
Reconstruction of the 3D model of AMPK trimer in basal state
Method: single particle / : Zhu L, Chen L, Zhou XM, Zhang YY, Zhang YJ, Zhao J, Ji SR, Wu JW, Wu Y
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
