-Search query
-Search result
Showing 1 - 50 of 218 items for (author: sergey & n)
EMDB-44479: 
Cryo-EM structure of synthetic claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and Nanobody

PDB-9bei: 
Cryo-EM structure of synthetic claudin-4 complex with Clostridium perfringens enterotoxin C-terminal domain, sFab COP-2, and Nanobody

EMDB-19067: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).

EMDB-19076: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).

EMDB-19077: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).

PDB-8rd8: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1).

PDB-8rdv: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2).

PDB-8rdw: 
Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3).
EMDB-43074: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)
EMDB-43075: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)
EMDB-43076: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)

EMDB-43077: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Structure 6)

EMDB-43078: 
Hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) bound to Mycobacterium smegmatis 70S ribosome, from focused 3D classification and refinement (Structure 6)

PDB-8v9j: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4)

PDB-8v9k: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Rv2629 (Balon) (Structure 5)

PDB-8v9l: 
Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) and MsmegEF-Tu(GDP) (Composite structure 6)
EMDB-42455: 
Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome

PDB-8upt: 
Candidatus Methanomethylophilus alvus tRNAPyl in A-site of ribosome

EMDB-18874: 
Cofactor-free Tau 4R2N isoform

PDB-8r3t: 
Cofactor-free Tau 4R2N isoform

EMDB-17402: 
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus

EMDB-18415: 
Cysteine tRNA ligase homodimer

PDB-8p49: 
Uncharacterized Q8U0N8 protein from Pyrococcus furiosus

PDB-8qhp: 
Cysteine tRNA ligase homodimer

EMDB-40675: 
Cryogenic electron microscopy map of human plakophilin-3
EMDB-40557: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)

PDB-8sk7: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
EMDB-26830: 
CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex

EMDB-26832: 
CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate

PDB-7uwe: 
CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex

PDB-7uwh: 
CryoEM Structure of E. coli Transcription-Coupled Ribonucleotide Excision Repair (TC-RER) complex bound to ribonucleotide substrate

EMDB-29491: 
CryoEM structure of E.coli transcription elongation complex

EMDB-29494: 
CryoEM structure of E.coli transcription elongation complex bound to ppGpp

PDB-8fvr: 
CryoEM structure of E.coli transcription elongation complex

PDB-8fvw: 
CryoEM structure of E.coli transcription elongation complex bound to ppGpp

EMDB-13795: 
Cryo-EM structure of TDP43 core peptide amyloid fiber

PDB-7q3u: 
Cryo-EM structure of TDP43 core peptide amyloid fiber
EMDB-15592: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqw: 
BA.4/5 SARS-CoV-2 Spike bound to mouse ACE2 (local)
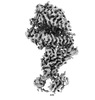
EMDB-15588: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local)

EMDB-15589: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (local)

EMDB-15590: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
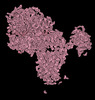
EMDB-15591: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqs: 
BA.4/5 SARS-CoV-2 Spike bound to human ACE2 (local)

PDB-8aqt: 
Beta SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqu: 
BA.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)

PDB-8aqv: 
BA.2.12.1 SARS-CoV-2 Spike bound to mouse ACE2 (local)
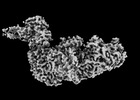
EMDB-14847: 
Cryo-EM structure of a CRISPR effector in complex with regulator

EMDB-14848: 
Cryo-EM structure of a CRISPR effector in complex with a caspase regulator

PDB-7zol: 
Cryo-EM structure of a CRISPR effector in complex with regulator
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
