+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryogenic electron microscopy map of human plakophilin-3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | actin cytoskeleton / armadillo / desmosomes / phosphatidylinositol 4 / 5-bisphosphate / plasma membrane / plakophilin / CELL ADHESION | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
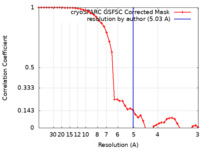
| Method | single particle reconstruction / cryo EM / Resolution: 5.03 Å | |||||||||
 Authors Authors | Gupta J / Izard T / Rangarajan ES | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Int J Mol Sci / Year: 2023 Journal: Int J Mol Sci / Year: 2023Title: Plakophilin-3 Binds the Membrane and Filamentous Actin without Bundling F-Actin. Authors: Jyoti Gupta / Erumbi S Rangarajan / Regina B Troyanovsky / Indrajyoti Indra / Sergey M Troyanovsky / Tina Izard /  Abstract: Plakophilin-3 is a ubiquitously expressed protein found widely in epithelial cells and is a critical component of desmosomes. The plakophilin-3 carboxy-terminal domain harbors nine armadillo repeat ...Plakophilin-3 is a ubiquitously expressed protein found widely in epithelial cells and is a critical component of desmosomes. The plakophilin-3 carboxy-terminal domain harbors nine armadillo repeat motifs with largely unknown functions. Here, we report the 5 Å cryogenic electron microscopy (cryoEM) structure of the armadillo repeat motif domain of plakophilin-3, one of the smaller cryoEM structures reported to date. We find that this domain is a monomer or homodimer in solution. In addition, using an in vitro actin co-sedimentation assay, we show that the armadillo repeat domain of plakophilin-3 directly interacts with F-actin. This feature, through direct interactions with actin filaments, could be responsible for the observed association of extra-desmosomal plakophilin-3 with the actin cytoskeleton directly attached to the adherens junctions in A431 epithelial cells. Further, we demonstrate, through lipid binding analyses, that plakophilin-3 can effectively be recruited to the plasma membrane through phosphatidylinositol-4,5-bisphosphate-mediated interactions. Collectively, we report on novel properties of plakophilin-3, which may be conserved throughout the plakophilin protein family and may be behind the roles of these proteins in cell-cell adhesion. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40675.map.gz emd_40675.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40675-v30.xml emd-40675-v30.xml emd-40675.xml emd-40675.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40675_fsc.xml emd_40675_fsc.xml | 4.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_40675.png emd_40675.png | 34.2 KB | ||
| Others |  emd_40675_half_map_1.map.gz emd_40675_half_map_1.map.gz emd_40675_half_map_2.map.gz emd_40675_half_map_2.map.gz | 7.4 MB 7.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40675 http://ftp.pdbj.org/pub/emdb/structures/EMD-40675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40675 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40675 | HTTPS FTP |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40675.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40675.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.44 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40675_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
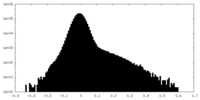
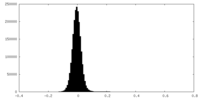
| Density Histograms |
-Half map: #2
| File | emd_40675_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
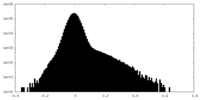
| Density Histograms |
- Sample components
Sample components
-Entire : plakophilin-3
| Entire | Name: plakophilin-3 |
|---|---|
| Components |
|
-Supramolecule #1: plakophilin-3
| Supramolecule | Name: plakophilin-3 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 57.2 KDa |
-Macromolecule #1: plakophilin-3
| Macromolecule | Name: plakophilin-3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: HHHHHHHHSS GLEVLFQGPG SHRTLQ RLS SGFDDID LP SAVKYLMA S DPNLQVLGA AYIQHKCYSD AAAKKQARS L QAVPRLVK LF NHANQEV QRH ATGAMR NLIY DNADN KLALV EENG IFELLR TLR EQDDELR KN VTGILWNL S SSDHLKDRL ...String: HHHHHHHHSS GLEVLFQGPG SHRTLQ RLS SGFDDID LP SAVKYLMA S DPNLQVLGA AYIQHKCYSD AAAKKQARS L QAVPRLVK LF NHANQEV QRH ATGAMR NLIY DNADN KLALV EENG IFELLR TLR EQDDELR KN VTGILWNL S SSDHLKDRL ARDTLEQLTD LVLSPLSGA G GPPLIQQN AS EAEIFYN ATG FLRNLS SASQ ATRQK MRECH GLVD ALVTSI NHA LDAGKCE DK SVENAVCV L RNLSYRLYD EMPPSALQRL EGRGRRDLA G APPGEVVG CF TPQSRRL REL PLAADA LTFA EVSKD PKGLE WLWS PQIVGL YNR LLQRCEL NR HTTEAAAG A LQNITAGDR RWAGVLSRLA LEQERILNP L LDRVRTAD HH QLRSLTG LIR NLSRNA RNKD EMSTK VVSHL IEKL PGSVGE KSP PAEVLVN II AVLNNLVV A SPIAARDLL YFDGLRKLIF IKKKRDSPD S EKSSRAAS SL LANLWQY NKL HRDFRA KGYR KEDFL GP |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: 20 mM Tris, 150 mM NaCl, 1 mM DTT |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 180 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.04 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 281 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)