-検索条件
-検索結果
検索 (著者・登録者: mace & k)の結果59件中、1から50件目までを表示しています

EMDB-19478: 
O-Layer C14 at 2.46A - Local refinement with C14 symmetry of the O-layer of the outer membrane core complex from the fully-assembled R388 type IV secretion system.

EMDB-19479: 
I-Layer C16 at 2.69A - Local refinement with C16 symmetry of the I-layer of the outer membrane core complex from the fully-assembled R388 type IV secretion system.
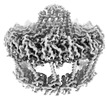
EMDB-19480: 
Conformation-A OMCC at 3.18A - Refinement without symmetry of the outer membrane core complex from the fully-assembled R388 type IV secretion system.
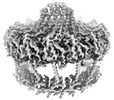
EMDB-19481: 
Conformation-B of the full-length outer membrane core complex (TrwH/VirB7, TrwF/VirB9, TrwE/VirB10CTD) from the fully-assembled R388 type IV secretion system determined by cryo-EM.
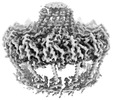
EMDB-19482: 
Conformation-C OMCC at 3.05A - Refinement without symmetry of the outer membrane core complex from the fully-assembled R388 type IV secretion system.

EMDB-19483: 
Stalk C5 at 2.97A - Local refinement with C5 symmetry of the Stalk complex from the fully-assembled R388 type IV secretion system.

EMDB-19484: 
Arches-protomer at 6.22A - Refinement without symmetry of the Arches-protomer complex from the fully-assembled R388 type IV secretion system.

EMDB-19485: 
Extended inner membrne complex (IMC) protomer at 3.83A - Refinement without symmetry of the extended IMC-protomer complex from the fully-assembled R388 type IV secretion system.

EMDB-19488: 
Stalk-Arches-IMC at 4.33A - Refinement without symmetry of the Stalk-Arches-IMC from the fully-assembled R388 type IV secretion.

PDB-8rt4: 
O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rt5: 
I-layer structure (TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rt6: 
Conformation-A of the full-length outer membrane core complex (TrwH/VirB7, TrwF/VirB9, TrwE/VirB10CTD) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rt7: 
Conformation-B of the full-length outer membrane core complex (TrwH/VirB7, TrwF/VirB9, TrwE/VirB10CTD) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rt8: 
Conformation-C of the full-length outer membrane core complex (TrwH/VirB7, TrwF/VirB9, TrwE/VirB10CTD) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rt9: 
Stalk complex full-length structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rta: 
Arches-protomer complex full-length structure (TrwJ/VirB8) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rtb: 
Extended inner membrane complex (IMC) protomer structure (TrwM/VirB3-TrwK/VirB4-TrwI/VirB6-TrwG/VirB8-TrwE/VirB10) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-8rtd: 
Stalk-Arches-IMC structure from the fully-assembled R388 type IV secretion system determined by cryo-EM.

EMDB-41265: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer)

EMDB-41266: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer)

PDB-8thi: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (parallel dimer)

PDB-8thj: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Haemophilus influenzae (antiparallel dimer)

EMDB-15775: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc

PDB-8b01: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in a nanodisc

EMDB-13968: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol

PDB-7qha: 
Cryo-EM structure of the Tripartite ATP-independent Periplasmic (TRAP) transporter SiaQM from Photobacterium profundum in amphipol

EMDB-12707: 
O-Layer C14 at 2.58A - Local refinement with C14 symmetry of the O-layer of the outer membrane core complex from the fully-assembled R388 type IV secretion system.

EMDB-12708: 
I-Layer C16 at 3.08A - Local refinement with C16 symmetry of I-layer of the outer membrane core complex from the fully-assembled R388 type IV secretion system.

EMDB-12709: 
Stalk C1 at 3.71A - Local refinement without symmetry of the Stalk complex from the fully-assembled R388 type IV secretion system.

EMDB-12715: 
IMC-Arches C6 at 8.33A - Refinement with C6 symmetry of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system.
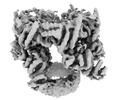
EMDB-12716: 
Trimer of dimers TrwK/VirB4unbound C1 at 4.14A - Refinement without symmetry of TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system.

EMDB-12717: 
Dimer TrwK/VirB4unbound C1 at 3.49A - Local refinement without symmetry of the TrwK/VirB4unbound dimer complex from the R388 type IV secretion system.

EMDB-12933: 
IMC protomer C1 at 3.75A - Local refinement without symmetry of the inner membrane complex (IMC) protomer from the fully-assembled R388 type IV secretion system.

EMDB-13765: 
OMCC C1 at 3.28A - Refinement without symmetry of the outer membrane core complex (O- and I-layer) from the fully-assembled R388 type IV secretion system.

EMDB-13766: 
Ab Initio model for IMC-Arches-Stalk from the fully-assembled R388 type IV secretion system determined by cryo-EM.

EMDB-13767: 
IMC-Arches-Stalk C1 at 6.18A - Refinement of the IMC, Arches and Stalk complex without symmetry from the fully-assembled R388 type IV secretion system.

EMDB-13768: 
Stalk C5 at 3.28A - Local refinement with C5 symmetry of the Stalk complex from the fully-assembled R388 type IV secretion system.

PDB-7o3j: 
O-layer structure (TrwH/VirB7, TrwF/VirB9CTD, TrwE/VirB10CTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-7o3t: 
I-layer structure (TrwF/VirB9NTD, TrwE/VirB10NTD) of the outer membrane core complex from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-7o3v: 
Stalk complex structure (TrwJ/VirB5-TrwI/VirB6) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-7o41: 
Hexameric composite model of the Inner Membrane Complex (IMC) with the Arches from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-7o42: 
TrwK/VirB4unbound trimer of dimers complex (with Hcp1) from the R388 type IV secretion system determined by cryo-EM.

PDB-7o43: 
TrwK/VirB4unbound dimer complex from R388 type IV secretion system determined by cryo-EM.

PDB-7oiu: 
Inner Membrane Complex (IMC) protomer structure (TrwM/VirB3, TrwK/VirB4, TrwG/VirB8tails) from the fully-assembled R388 type IV secretion system determined by cryo-EM.

PDB-7q1v: 
Arches protomer (trimer of TrwG/VirB8peri) structure from the fully-assembled R388 type IV secretion system determined by cryo-EM.

EMDB-13083: 
L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains

EMDB-13858: 
L. pneumophila Type IV Coupling Complex (T4CC) computed from WT particles where DotY/Z is missing (referred to as T4CCWTminusYZ at 6.3A)

EMDB-13859: 
L. pneumophila Type IV Coupling Complex (T4CC) computed from a sample where dotY/Z genes were deleted (referred to as T4CCdeltaDotYZ at 15A)

PDB-7ovb: 
L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains

EMDB-10350: 
Type IV Coupling Complex (T4CC) from L. pneumophila.
ページ:
 ムービー
ムービー コントローラー
コントローラー 構造ビューア
構造ビューア EMN検索について
EMN検索について



 wwPDBはEMDBデータモデルのバージョン3へ移行します
wwPDBはEMDBデータモデルのバージョン3へ移行します
