[English] 日本語
 Yorodumi
Yorodumi- EMDB-13766: Ab Initio model for IMC-Arches-Stalk from the fully-assembled R38... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ab Initio model for IMC-Arches-Stalk from the fully-assembled R388 type IV secretion system determined by cryo-EM. | |||||||||||||||
 Map data Map data | Initial model for IMC-Arches-Stalk complex - Refinement without symmetry sharpened | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | type IV secretion system / type 4 secretion system / T4SS / IMC / inner membrane complex / inner membrane / arches / stalk / periplasmic / R388 plasmid / conjugation / bacterial secretion / secretion / secretion system / protein complex / VirB3 / VirB4 / VirB5 / VirB6 / VirB8 / TrwM / TrwK / TrwJ / TrwI / TrwG / MEMBRANE PROTEIN | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 9.2 Å | |||||||||||||||
 Authors Authors | Mace K / Vadakkepat AK / Waksman G / Lukoyanova N | |||||||||||||||
| Funding support |  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Cryo-EM structure of a type IV secretion system. Authors: Kévin Macé / Abhinav K Vadakkepat / Adam Redzej / Natalya Lukoyanova / Clasien Oomen / Nathalie Braun / Marta Ukleja / Fang Lu / Tiago R D Costa / Elena V Orlova / David Baker / Qian Cong ...Authors: Kévin Macé / Abhinav K Vadakkepat / Adam Redzej / Natalya Lukoyanova / Clasien Oomen / Nathalie Braun / Marta Ukleja / Fang Lu / Tiago R D Costa / Elena V Orlova / David Baker / Qian Cong / Gabriel Waksman /     Abstract: Bacterial conjugation is the fundamental process of unidirectional transfer of DNAs, often plasmid DNAs, from a donor cell to a recipient cell. It is the primary means by which antibiotic resistance ...Bacterial conjugation is the fundamental process of unidirectional transfer of DNAs, often plasmid DNAs, from a donor cell to a recipient cell. It is the primary means by which antibiotic resistance genes spread among bacterial populations. In Gram-negative bacteria, conjugation is mediated by a large transport apparatus-the conjugative type IV secretion system (T4SS)-produced by the donor cell and embedded in both its outer and inner membranes. The T4SS also elaborates a long extracellular filament-the conjugative pilus-that is essential for DNA transfer. Here we present a high-resolution cryo-electron microscopy (cryo-EM) structure of a 2.8 megadalton T4SS complex composed of 92 polypeptides representing 8 of the 10 essential T4SS components involved in pilus biogenesis. We added the two remaining components to the structural model using co-evolution analysis of protein interfaces, to enable the reconstitution of the entire system including the pilus. This structure describes the exceptionally large protein-protein interaction network required to assemble the many components that constitute a T4SS and provides insights on the unique mechanism by which they elaborate pili. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13766.map.gz emd_13766.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13766-v30.xml emd-13766-v30.xml emd-13766.xml emd-13766.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_13766.png emd_13766.png | 63.1 KB | ||
| Masks |  emd_13766_msk_1.map emd_13766_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13766.cif.gz emd-13766.cif.gz | 4.2 KB | ||
| Others |  emd_13766_additional_1.map.gz emd_13766_additional_1.map.gz emd_13766_half_map_1.map.gz emd_13766_half_map_1.map.gz emd_13766_half_map_2.map.gz emd_13766_half_map_2.map.gz | 31.6 MB 59.5 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13766 http://ftp.pdbj.org/pub/emdb/structures/EMD-13766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13766 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13766 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13766.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13766.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Initial model for IMC-Arches-Stalk complex - Refinement without symmetry sharpened | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.134 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13766_msk_1.map emd_13766_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
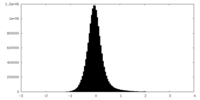
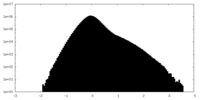
| Density Histograms |
-Additional map: Initial model for IMC-Arches-Stalk complex - Refinement without...
| File | emd_13766_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Initial model for IMC-Arches-Stalk complex - Refinement without symmetry unsharpened | ||||||||||||
| Projections & Slices |
| ||||||||||||
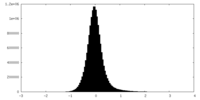
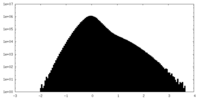
| Density Histograms |
-Half map: Initial model for IMC-Arches-Stalk complex - Half-A
| File | emd_13766_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Initial model for IMC-Arches-Stalk complex - Half-A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Initial model for IMC-Arches-Stalk complex - Half-B
| File | emd_13766_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Initial model for IMC-Arches-Stalk complex - Half-B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Type IV secretion system complex
| Entire | Name: Type IV secretion system complex |
|---|---|
| Components |
|
-Supramolecule #1: Type IV secretion system complex
| Supramolecule | Name: Type IV secretion system complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.808 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 57.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER / Details: CryoSPARC ab-initio |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 9.2 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 28902 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: OTHER / Details: Stochastic gradient descent (SGD) |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)