-Search query
-Search result
Showing 1 - 50 of 566 items for (author: ma & yb)

EMDB-44482: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab
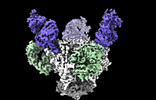
EMDB-44484: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs
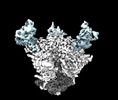
EMDB-44491: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

PDB-9ber: 
Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with PGT122 Fab

PDB-9bew: 
Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with 3BNC117 and 10-1074 Fabs

PDB-9bf6: 
Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with PGT122 Fab

EMDB-29620: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B)
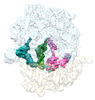
EMDB-29621: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A)
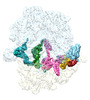
EMDB-29627: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A)
EMDB-29628: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D)

EMDB-29631: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B)
EMDB-29634: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C)

PDB-8fzd: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with apoRF3, RF1, P- and E-site tRNAPhe (Composite state I-B)

PDB-8fze: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State I-A)

PDB-8fzg: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF3-GDPCP, RF1, P- and E-site tRNAPhe (Composite state II-A)

PDB-8fzh: 
Cryo-EM structure of an E. coli non-rotated ribosome termination complex bound with RF1, P- and E-site tRNAPhe (State II-D)

PDB-8fzi: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-B)

PDB-8fzj: 
Cryo-EM structure of an E. coli rotated ribosome bound with RF3-GDPCP and p/E-tRNAPhe (Composite state II-C)
EMDB-29622: 
Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C)
EMDB-29623: 
RF3-ppGpp bound to an E. coli rotated ribosome, from focused classification and refinement (State I-C)
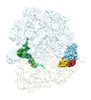
EMDB-29624: 
Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C)

PDB-8fzf: 
Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (Composite state I-C)

EMDB-18723: 
Cryo-EM structure of the cross-exon pre-B complex (tri-snRNP region)

EMDB-18724: 
Cryo-EM structure of the cross-exon pre-B+5'ss complex (tri-snRNP region)

EMDB-18725: 
Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex(tri-snRNP region)

EMDB-18726: 
Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPgammaS complex (tri-snRNP region)

EMDB-18727: 
Cryo-EM structure of the cross-exon pre-B+AMPPNP complex (tri-snRNP region)
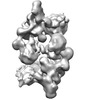
EMDB-19594: 
cryo-EM structure of dimerized cross-exon pre-B complex

EMDB-19595: 
cryo-EM structure of dimerized cross-exon B-like complex
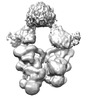
EMDB-19596: 
cryo-EM structure of dimerized cross-exon pre-B+5'ss+ATPyS complex

EMDB-19597: 
cryo-EM structure of dimerized cross-exon pre-B+5'ssLNG+ATPyS complex

EMDB-19598: 
cryo-EM structure of dimerized cross-exon pre-B+ATP complex
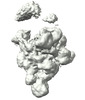
EMDB-19847: 
cryo-EM structure of pre-B+5'ss complex (incubated at 30 degree)
EMDB-19848: 
cryo-EM structure of cross-exon pre-B+5'ss+ATP complex

EMDB-19868: 
cryo-EM structure of dimerized pre-B+5'ss complex

EMDB-18542: 
Cryo-EM Structure of Pre-B+5'ss+ATPgammaS Complex (core part)

EMDB-18544: 
Cryo-EM Structure of Pre-B Complex (core part)

EMDB-18545: 
Cryo-EM Structure of Pre-B+AMPPNP Complex (core part)

EMDB-18546: 
Cryo-EM Structure of Pre-B+5'ssLNG Complex (core part)

EMDB-18547: 
Cryo-EM Structure of Pre-B+ATP Complex (core part)

EMDB-18548: 
Cryo-EM Structure of Pre-B-like Complex (core part)

EMDB-18555: 
Cryo-EM Structure of Pre-B+5'ss Complex (core part)

EMDB-18718: 
Cryo-EM structure of the cross-exon pre-B complex

EMDB-18781: 
Cryo-EM structure of the cross-exon B-like complex

EMDB-18786: 
Cryo-EM structure of the cross-exon pre-B+AMPPNP complex

EMDB-18787: 
Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex

EMDB-18788: 
Cryo-EM structure of the cross-exon pre-B+5'ss complex

EMDB-18789: 
Cryo-EM structure of the cross-exon pre-B+ATP complex

EMDB-19349: 
Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPyS complex

PDB-8qoz: 
Cryo-EM Structure of Pre-B+5'ss+ATPgammaS Complex (core part)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
