[English] 日本語
 Yorodumi
Yorodumi- EMDB-29622: Cryo-EM structure of an E. coli rotated ribosome complex bound wi... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C) | ||||||||||||
 Map data Map data | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C) | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | release factor 3 / termination complex / cryo-EM / tRNA / RIBOSOME | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | ||||||||||||
 Authors Authors | Rybak MY / Li L / Lin J / Gagnon MG | ||||||||||||
| Funding support |  United States, United States,  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: The ribosome termination complex remodels release factor RF3 and ejects GDP. Authors: Li Li / Mariia Yu Rybak / Jinzhong Lin / Matthieu G Gagnon /   Abstract: Translation termination involves release factors RF1, RF2 and the GTPase RF3 that recycles RF1 and RF2 from the ribosome. RF3 dissociates from the ribosome in the GDP-bound form and must then ...Translation termination involves release factors RF1, RF2 and the GTPase RF3 that recycles RF1 and RF2 from the ribosome. RF3 dissociates from the ribosome in the GDP-bound form and must then exchange GDP for GTP. The 70S ribosome termination complex (70S-TC) accelerates GDP exchange in RF3, suggesting that the 70S-TC can function as the guanine nucleotide exchange factor for RF3. Here, we use cryogenic-electron microscopy to elucidate the mechanism of GDP dissociation from RF3 catalyzed by the Escherichia coli 70S-TC. The non-rotated ribosome bound to RF1 remodels RF3 and induces a peptide flip in the phosphate-binding loop, efficiently ejecting GDP. Binding of GTP allows RF3 to dock at the GTPase center, promoting the dissociation of RF1 from the ribosome. The structures recapitulate the functional cycle of RF3 on the ribosome and uncover the mechanism by which the 70S-TC allosterically dismantles the phosphate-binding groove in RF3, a previously overlooked function of the ribosome. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_29622.map.gz emd_29622.map.gz | 255.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-29622-v30.xml emd-29622-v30.xml emd-29622.xml emd-29622.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_29622.png emd_29622.png | 154 KB | ||
| Masks |  emd_29622_msk_1.map emd_29622_msk_1.map | 512 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-29622.cif.gz emd-29622.cif.gz | 4.4 KB | ||
| Others |  emd_29622_additional_1.map.gz emd_29622_additional_1.map.gz emd_29622_half_map_1.map.gz emd_29622_half_map_1.map.gz emd_29622_half_map_2.map.gz emd_29622_half_map_2.map.gz | 444.9 MB 474.9 MB 474.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-29622 http://ftp.pdbj.org/pub/emdb/structures/EMD-29622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29622 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-29622 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_29622.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_29622.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||
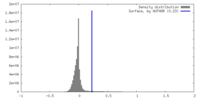
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_29622_msk_1.map emd_29622_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
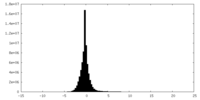
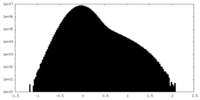
| Density Histograms |
-Additional map: Cryo-EM structure of an E. coli rotated ribosome...
| File | emd_29622_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C). Sharpened map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
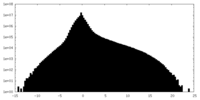
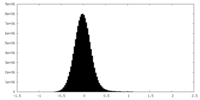
| Density Histograms |
-Half map: Cryo-EM structure of an E. coli rotated ribosome...
| File | emd_29622_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C). Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
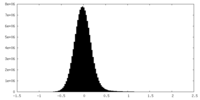
| Density Histograms |
-Half map: Cryo-EM structure of an E. coli rotated ribosome...
| File | emd_29622_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C). Half map. | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of an E. coli rotated ribosome complex bound wi...
| Entire | Name: Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C) |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of an E. coli rotated ribosome complex bound wi...
| Supramolecule | Name: Cryo-EM structure of an E. coli rotated ribosome complex bound with RF3-ppGpp and p/E-tRNAPhe (State I-C) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.6 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Details: 5 mM Tris-HCl, 60 mM NH4Cl, 10 mM MgCl2, 6 mM B-mercaptoethanol |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: GOLD / Mesh: 200 / Support film - Material: GOLD / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 295 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: INTEGRATING / Number grids imaged: 1 / Number real images: 10284 / Average exposure time: 1.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)