-Search query
-Search result
Showing all 42 items for (author: kumar & yd)

EMDB-74033: 
1-methyl-pseudouridine twist corrected RNA origami 6-helix bundle type-1 dimer
Method: single particle / : McRae EKS, Kumar YD

EMDB-44199: 
Biased agonist bound CB1-Gi structure
Method: single particle / : Rangari VA, O'Brien ES, Kobilka BK, Krishna Kumar K, Majumdar S

EMDB-44247: 
Biased agonist bound CB1-Gi structure
Method: single particle / : Rangari VA, O'Brien ES, Kobilka BK, Krishna Kumar K, Majumdar S

PDB-9b54: 
Biased agonist bound CB1-Gi structure
Method: single particle / : Rangari VA, O'Brien ES, Kobilka BK, Krishna Kumar K, Majumdar S

PDB-9b65: 
Biased agonist bound CB1-Gi structure
Method: single particle / : Rangari VA, O'Brien ES, Kobilka BK, Krishna Kumar K, Majumdar S

EMDB-51640: 
Subtomogram average of immature Langat virus from cryo-electron tomograms of infected cells
Method: subtomogram averaging / : Carlson LA, Dahmane S

EMDB-51642: 
Subtomogram average of mature Langat virus
Method: subtomogram averaging / : Carlson LA, Dahmane S

EMDB-18778: 
Structure of DNMT3A1 UDR region bound to H2AK119ub nucleosome
Method: single particle / : Wapenaar H, Wilson MD

EMDB-18793: 
Cryo-EM density map of DNMT3A1-DNMT3L on a human H2AKc119ub nucleosome at 5.1 A resolution
Method: single particle / : Wapenaar H, Wilson MD

PDB-8qzm: 
Structure of DNMT3A1 UDR region bound to H2AK119ub nucleosome
Method: single particle / : Burdett H, Wapenaar H, Wilson MD

EMDB-42074: 
Representative tomogram of Enterococcus faecium WT Com15
Method: electron tomography / : Hang HC, Park D

EMDB-42086: 
Representative tomogram of Enterococcus faecium SagA complementation strain
Method: electron tomography / : Hang HC, Park D

EMDB-42087: 
Representative tomogram of Enterococcus faecium SagA deletion strain
Method: electron tomography / : Hang HC, Park D

EMDB-16859: 
Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes (Map1)
Method: single particle / : Foglizzo M, Burdett H, Wilson MD, Zeqiraj E

EMDB-16869: 
Low-resolution structure of BRCA1dExon11-FL BARD1 (closed state)
Method: single particle / : Foglizzo M, Zeqiraj E

EMDB-16870: 
Low-resolution structure of BRCA1dExon11-FL BARD1 (open state)
Method: single particle / : Foglizzo M, Zeqiraj E

EMDB-17928: 
Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes (Map 2)
Method: single particle / : Foglizzo M, Burdett H, Wilson MD, Zeqiraj E

PDB-8off: 
Structure of BARD1 ARD-BRCTs in complex with H2AKc15ub nucleosomes (Map1)
Method: single particle / : Foglizzo M, Burdett H, Wilson MD, Zeqiraj E
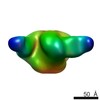
EMDB-8078: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.12 wk323 Fab
Method: single particle / : Ozorowski G, Ward AB

EMDB-8079: 
B41 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.12 wk323 Fab
Method: single particle / : Ozorowski G, Ward AB
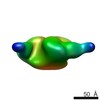
EMDB-8080: 
B41 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.9 wk152 Fab
Method: single particle / : Ozorowski G, Ward AB

EMDB-8081: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235.9 wk152 Fab
Method: single particle / : Ozorowski G, Ward AB
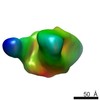
EMDB-8082: 
BG505 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235 wk41 Fab
Method: single particle / : Ozorowski G, Ward AB
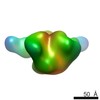
EMDB-8083: 
B41 SOSIP.664 HIV-1 Env trimer in complex with anti-HIV CH235 wk41 Fab
Method: single particle / : Ozorowski G, Ward AB

PDB-3izh: 
Mm-cpn D386A with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3izi: 
Mm-cpn rls with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3izj: 
Mm-cpn rls with ATP and AlFx
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3izk: 
Mm-cpn rls deltalid with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3izl: 
Mm-cpn rls deltalid with ATP and AlFx
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3izm: 
Mm-cpn wildtype with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3izn: 
Mm-cpn deltalid with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5244: 
Mm-cpn D386A with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5245: 
Mm-cpn rls with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5246: 
Mm-cpn rls with ATP and AlFx
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5247: 
Mm-cpn rls deltalid with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5248: 
Mm-cpn rls deltalid with ATP and AlFx
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5249: 
Mm-cpn wildtype with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

EMDB-5250: 
Mm-cpn deltalid with ATP
Method: single particle / : Douglas NR, Reissmann S, Zhang J, Chen B, Jakana J, Kumar R, Chiu W, Frydman J

PDB-3iyg: 
Ca model of bovine TRiC/CCT derived from a 4.0 Angstrom cryo-EM map
Method: single particle / : Cong Y, Baker ML, Ludtke SJ, Frydman J, Chiu W

PDB-3ktt: 
Atomic model of bovine TRiC CCT2(beta) subunit derived from a 4.0 Angstrom cryo-EM map
Method: single particle / : Cong Y, Baker ML, Ludtke SJ, Frydman J, Chiu W

EMDB-5145: 
4.7 Angstrom asymmetric cryo-EM map of TRiC in the both-ring closed ATP-AlFx state
Method: single particle / : Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W

EMDB-5148: 
4.0 Angstrom two-fold imposed cryo-EM map of TRiC in the both-ring closed ATP-AlFx state
Method: single particle / : Cong Y, Baker ML, Jakana J, Woolford D, Miller EJ, Reissmann S, Kumar RN, Redding-Johanson AM, Batth TS, Mukhopadhyay A, Ludtke SJ, Frydman J, Chiu W
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
