+Search query
-Structure paper
| Title | A cryptic pocket in CB1 drives peripheral and functional selectivity. |
|---|---|
| Journal, issue, pages | Nature, Vol. 640, Issue 8057, Page 265-273, Year 2025 |
| Publish date | Mar 5, 2025 |
 Authors Authors | Vipin Ashok Rangari / Evan S O'Brien / Alexander S Powers / Richard A Slivicki / Zachariah Bertels / Kevin Appourchaux / Deniz Aydin / Nokomis Ramos-Gonzalez / Juliet Mwirigi / Li Lin / Elizaveta Mangutov / Briana L Sobecks / Yaseen Awad-Agbaria / Manoj B Uphade / Jhoan Aguilar / Teja Nikhil Peddada / Yuki Shiimura / Xi-Ping Huang / Jakayla Folarin-Hines / Maria Payne / Anirudh Kalathil / Balazs R Varga / Brian K Kobilka / Amynah A Pradhan / Michael D Cameron / Kaavya Krishna Kumar / Ron O Dror / Robert W Gereau / Susruta Majumdar /   |
| PubMed Abstract | The current opioid overdose epidemic highlights the urgent need to develop safer and more effective treatments for chronic pain. Cannabinoid receptor type 1 (CB1) is a promising non-opioid target for ...The current opioid overdose epidemic highlights the urgent need to develop safer and more effective treatments for chronic pain. Cannabinoid receptor type 1 (CB1) is a promising non-opioid target for pain relief, but its clinical use has been limited by centrally mediated psychoactivity and tolerance. We overcame both issues by designing peripherally restricted CB1 agonists that minimize arrestin recruitment. We achieved these goals by computationally designing positively charged derivatives of the potent CB1 agonist MDMB-Fubinaca. We designed these ligands to occupy a cryptic pocket identified through molecular dynamics simulations-an extended binding pocket that opens rarely and leads to the conserved signalling residue D (ref. ). We used structure determination, pharmacological assays and molecular dynamics simulations to verify the binding modes of these ligands and to determine the molecular mechanism by which they achieve this dampening of arrestin recruitment. Our lead ligand, VIP36, is highly peripherally restricted and demonstrates notable efficacy in three mouse pain models, with 100-fold dose separation between analgesic efficacy and centrally mediated side effects. VIP36 exerts analgesic efficacy through peripheral CB1 receptors and shows limited analgesic tolerance. These results show how targeting a cryptic pocket in a G-protein-coupled receptor can lead to enhanced peripheral selectivity, biased signalling, desired in vivo pharmacology and reduced adverse effects. This has substantial implications for chronic pain treatment but could also revolutionize the design of drugs targeting other G-protein-coupled receptors. |
 External links External links |  Nature / Nature /  PubMed:40044849 / PubMed:40044849 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.86 - 3.03 Å |
| Structure data | EMDB-44199, PDB-9b54: EMDB-44247, PDB-9b65: |
| Chemicals |  PDB-1aiw: 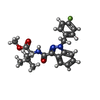 ChemComp-KCA: |
| Source |
|
 Keywords Keywords | SIGNALING PROTEIN / G Protein-coupled receptor (GPCR) / G protein / Gi / biased ligand |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers







 homo sapiens (human)
homo sapiens (human)
