-Search query
-Search result
Showing 1 - 50 of 188 items for (author: kim & kh)
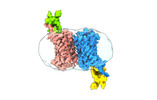
EMDB-36488: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-37212: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Receptor original map)
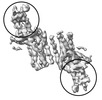
EMDB-37214: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Ligand/CCL7 focused map)

PDB-8jps: 
Structure of Duffy Antigen Receptor for Chemokines (DARC)/ACKR1 in complex with the chemokine, CCL7 (Composite map)

EMDB-43762: 
Aca2 from Pectobacterium phage ZF40 bound to RNA

PDB-8w35: 
Aca2 from Pectobacterium phage ZF40 bound to RNA

EMDB-42676: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

EMDB-42999: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

PDB-8uwl: 
5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM)

PDB-8v6u: 
5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking

EMDB-40825: 
10E8-GT10.2 immunogen in complex with human Fab 10E8 and mouse Fab W6-10

EMDB-43049: 
Cryo-electron tomogram of mitochondria from cryo-FIB milled l-Opa1* mouse embryonic fibroblasts

EMDB-43050: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts

EMDB-43051: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts

EMDB-43052: 
cryo-electron tomogram of mitochondria in cryo-FIB milled l-Opa1* mouse embryonic fibroblasts

EMDB-43053: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts

EMDB-43054: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts

EMDB-43055: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts

EMDB-43056: 
cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts

EMDB-43057: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled s-Opa1* mouse embryonic fibroblasts

EMDB-43058: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion

EMDB-43059: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion

EMDB-43060: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion

EMDB-43061: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion

EMDB-43062: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with OPA1 deletion

EMDB-43063: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts

EMDB-43064: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts
EMDB-43065: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts

EMDB-43066: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts

EMDB-43067: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1

EMDB-43068: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1

EMDB-43069: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1

EMDB-43070: 
Cryo-electron tomogram of mitochondria in cryo-FIB milled mouse embryonic fibroblasts with overexpression of OPA1

EMDB-41008: 
Cryo-EM structure of the DHA bound FFA4-Gq complex

EMDB-41010: 
Cryo-EM structure of the Butyrate bound FFA2-Gq complex

EMDB-41013: 
Cryo-EM structure of the DHA bound FFA1-Gq complex

PDB-8t3q: 
Cryo-EM structure of the DHA bound FFA4-Gq complex

PDB-8t3s: 
Cryo-EM structure of the Butyrate bound FFA2-Gq complex

PDB-8t3v: 
Cryo-EM structure of the DHA bound FFA1-Gq complex

EMDB-41007: 
Cryo-EM structure of the TUG-891 bound FFA4-Gq complex

EMDB-41014: 
Cryo-EM structure of the DHA bound FFA1-Gq complex(mask on receptor)

PDB-8t3o: 
Cryo-EM structure of the TUG-891 bound FFA4-Gq complex

EMDB-28156: 
Cryo-EM structure of human DNMT3B homo-tetramer (form I)

EMDB-28157: 
Cryo-EM structure of human DNMT3B homo-tetramer (form II)

EMDB-28158: 
Cryo-EM structure of human DNMT3B homo-trimer

EMDB-28159: 
Cryo-EM structure of human DNMT3B homo-hexamer

PDB-8eih: 
Cryo-EM structure of human DNMT3B homo-tetramer (form I)

PDB-8eii: 
Cryo-EM structure of human DNMT3B homo-tetramer (form II)

PDB-8eij: 
Cryo-EM structure of human DNMT3B homo-trimer

PDB-8eik: 
Cryo-EM structure of human DNMT3B homo-hexamer
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
