-Search query
-Search result
Showing all 39 items for (author: khan & mb)

EMDB-74113: 
Competition for different elements of the nucleosome acidic patch yields distinct functional outcomes. VHH 1B2
Method: single particle / : Chakraborty U, Saccone EC, Becerra GC, Khan LF, Arslanovic N, Aguilar R, Gloor SL, Hunt SR, Folkwein HJ, Husby NL, Maier KE, Marunde MR, Schomburg NK, Vaidya A, Cowles MW, Venters BJ, Kassavetis G, Sun ZW, Kadonaga JT, Armache JP, Keogh MC, Tyler JK

EMDB-74114: 
Competition for different elements of the nucleosome acidic patch yields distinct functional outcomes. VHH 1G1
Method: single particle / : Chakraborty U, Saccone EC, Becerra GC, Khan LF, Arslanovic N, Aguilar R, Gloor SL, Hunt SR, Folkwein HJ, Husby NL, Maier KE, Marunde MR, Schomburg NK, Vaidya A, Cowles MW, Venters BJ, Kassavetis G, Sun ZW, Kadonaga JT, Armache JP, Keogh MC, Tyler JK

PDB-9zen: 
Competition for different elements of the nucleosome acidic patch yields distinct functional outcomes. VHH 1B2
Method: single particle / : Chakraborty U, Saccone EC, Becerra GC, Khan LF, Arslanovic N, Aguilar R, Gloor SL, Hunt SR, Folkwein HJ, Husby NL, Maier KE, Marunde MR, Schomburg NK, Vaidya A, Cowles MW, Venters BJ, Kassavetis G, Sun ZW, Kadonaga JT, Armache JP, Keogh MC, Tyler JK

PDB-9zeo: 
Competition for different elements of the nucleosome acidic patch yields distinct functional outcomes. VHH 1G1
Method: single particle / : Chakraborty U, Saccone EC, Becerra GC, Khan LF, Arslanovic N, Aguilar R, Gloor SL, Hunt SR, Folkwein HJ, Husby NL, Maier KE, Marunde MR, Schomburg NK, Vaidya A, Cowles MW, Venters BJ, Kassavetis G, Sun ZW, Kadonaga JT, Armache JP, Keogh MC, Tyler JK

EMDB-48855: 
Cryo-EM structure of the magnesium transporter MgtA in E1-like and E2-P conformations at 2.59 angstroms
Method: single particle / : Khan MB, Primeau JO, Basu PC, Morth JP, Lemieux MJ, Young HS

EMDB-48923: 
Cryo-EM structure of the magnesium transporter MgtA in the E2 conformation with bound beryllium fluoride (BeF3-) and Mg2+ at 2.65 A resolution.
Method: single particle / : Khan MB, Primeau JO, Basu PC, Morth JP, Lemieux MJ, Young HS

EMDB-48602: 
Cryo-EM Structure of the Magnesium Transporter MgtA in the E2 Conformation Bound to Mg2+
Method: single particle / : Khan MB, Primeau JO, Basu PC, Morth JP, Lemieux MJ, Young HS

EMDB-48191: 
Cryo-EM structure of the magnesium transporter MgtA in an E1-like conformation with bound Mg2+ ions
Method: single particle / : Khan MB, Primeau JO, Basu PC, Morth JP, Lemieux MJ, Young HS

EMDB-45025: 
Cryo-EM structure of the magnesium transporter MgtA in the E2 conformation with bound lipids and Mg2+
Method: single particle / : Khan MB, Primeau JO, Young HS

EMDB-46892: 
Structure of SARS-CoV-2 spike in complex with antibody Fab COVIC-154
Method: single particle / : Yu X, Saphire EO

PDB-9dhy: 
Structure of SARS-CoV-2 spike in complex with antibody Fab COVIC-154
Method: single particle / : Yu X, Saphire EO

EMDB-25183: 
P. chlororaphis 70S ribosome in situ subtomogram average
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25220: 
In situ subtomogram average of the 201phi2-1 phage nucleus major shell protein, chimallin (concave class)
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25221: 
In situ consensus subtomogram average of the 201phi2-1 chimallin
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25222: 
In situ subtomogram average of 201phi2-1 phage nucleus major shell protein, chimallin (intermediate/flat class)
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25223: 
In situ subtomogram average of the 201phi2-1 phage nucleus major shell protein, chimallin (convex class)
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25229: 
In situ subtomogram average of the Goslar major phage nucleus shell protein, chimallin (consensus class)
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25262: 
In situ subtomogram average of Goslar phage nucleus major shell protein, chimallin (concave class)
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25358: 
In situ subtomogram average of the major Goslar phage nucleus shell protein, chimallin (convex class)
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25359: 
In situ subtomogram average of the APEC2248 70S ribosome
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25360: 
In situ subtomogram average of the APEC2248 50S ribosome
Method: subtomogram averaging / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25390: 
201Phi2-1 Chimallin Cubic (O, 24mer) assembly
Method: single particle / : Laughlin TG, Deep A

EMDB-25391: 
201phi2-1 Chimallin localized tetramer reconstruction
Method: single particle / : Laughlin TG, Deep A

EMDB-25392: 
201phi2-1 Chimallin C1 localized reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25393: 
201phi2-1 chimallin rectangular (D4,40mer) assembly
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25394: 
Goslar chimallin cubic (O, 24mer) assembly
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25395: 
Goslar chimallin C4 tetramer localized reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25396: 
Goslar chimallin C1 localized reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

PDB-7sqq: 
201Phi2-1 Chimallin Cubic (O, 24mer) assembly
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

PDB-7sqr: 
201phi2-1 Chimallin localized tetramer reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

PDB-7sqs: 
201phi2-1 Chimallin C1 localized reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

PDB-7sqt: 
Goslar chimallin cubic (O, 24mer) assembly
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

PDB-7squ: 
Goslar chimallin C4 tetramer localized reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

PDB-7sqv: 
Goslar chimallin C1 localized reconstruction
Method: single particle / : Laughlin TG, Deep A, Prichard AM, Seitz C, Gu Y, Enustun E, Suslov S, Khanna K, Birkholz EA, Amaro RE, Pogliano J, Corbett KD, Villa E

EMDB-25216: 
Slice of a cryo-electron tomogram of PhiPA3-infected Pseudomonas aeruginosa cell at 70 mpi (Cell 4)
Method: electron tomography / : Khanna K, Villa E

EMDB-25217: 
Slice of a cryo-electron tomogram of PhiPA3-infected Pseudomonas aeruginosa cell at 70 mpi (Cell 3)
Method: electron tomography / : Khanna K, Villa E

EMDB-25218: 
Slice of a cryo-electron tomogram of PhiPA3-infected Pseudomonas aeruginosa cell at 70 mpi (Cell 2)
Method: electron tomography / : Khanna K, Villa E

EMDB-25219: 
Slice of a cryo-electron tomogram of PhiPA3-infected Pseudomonas aeruginosa cell at 70 mpi showing phage bouquets (same as Fig 4A of the manuscript)
Method: electron tomography / : Khanna K, Villa E
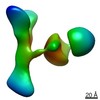
EMDB-25708: 
GPC2 HEP CT3 complex
Method: single particle / : Zhu J, Cachau R, De Val Alda N, Li N, Ho M
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
