-Search query
-Search result
Showing 1 - 50 of 1,057 items for (author: kang & g)

EMDB-37727: 
Cryo-ET structure of RuBisCO from 3.9 angstroms Synechococcus elongatus PCC 7942

EMDB-37728: 
Cryo-ET map of RuBisCO at 4.4 angstroms from Synechococcus elongatus PCC 7942 beta-carboxysome
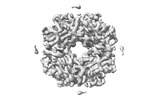
EMDB-37729: 
Cryo-ET map of RuBisCO-SSUL at 5.9 angstroms from Synechococcus elongatus PCC 7942 beta-carboxysome

EMDB-37730: 
Cryo-ET map of RuBisCO at the outermost layer that is loosely attached to the shell of Synechococcus elongatus PCC 7942 beta-carboxysome

EMDB-37731: 
Cryo-ET map of RuBisCO at the outermost layer that is tightly attached to the shell of Synechococcus elongatus PCC 7942 beta-carboxysome

PDB-8wpz: 
Cryo-ET structure of RuBisCO at 3.9 angstroms from Synechococcus elongatus PCC 7942

EMDB-36366: 
Cryo-EM structure of Symbiodinium photosystem I

PDB-8jjr: 
Cryo-EM structure of Symbiodinium photosystem I

EMDB-40751: 
Isobutyryl-CoA mutase fused Q341A in the presence of GTP

PDB-8ssl: 
Isobutyryl-CoA mutase fused Q341A in the presence of GTP

EMDB-36849: 
Nipah virus Attachment glycoprotein with 41-6 antibody fragment

PDB-8k3c: 
Nipah virus Attachment glycoprotein with 41-6 antibody fragment
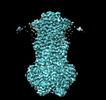
EMDB-40180: 
MsbA bound to cerastecin C

PDB-8gk7: 
MsbA bound to cerastecin C

EMDB-36228: 
Cryo-EM structure of Mi3 fused with FKBP

EMDB-36230: 
Cryo-EM structure of Mi3 fused with LOV2

PDB-8jga: 
Cryo-EM structure of Mi3 fused with FKBP

PDB-8jgc: 
Cryo-EM structure of Mi3 fused with LOV2

EMDB-37154: 
Cyanophage A-1(L) neck/gp7-terminator

EMDB-37155: 
Cyanophage A-1(L) neck/gp5-neck fiber

PDB-8kef: 
Cyanophage A-1(L) neck/gp7-terminator

PDB-8keg: 
Cyanophage A-1(L) neck/gp5-neck fiber

EMDB-37150: 
The CBD domain of cyanophage A-1(L) short tail fiber

EMDB-37151: 
Cyanophage A-1(L) baseplate-initiators

EMDB-37152: 
Cyanophage A-1(L) tail fiber

EMDB-37153: 
Cyanophage A-1(L) sheath-tube

EMDB-41590: 
Cyanophage A-1(L) portal

PDB-8ke9: 
The CBD domain of cyanophage A-1(L) short tail fiber

PDB-8kea: 
Cyanophage A-1(L) baseplate-initiators

PDB-8kec: 
Cyanophage A-1(L) tail fiber

PDB-8kee: 
Cyanophage A-1(L) sheath-tube

PDB-8ts6: 
Cyanophage A-1(L) portal

EMDB-40208: 
Backbone model of de novo-designed chlorophyll-binding nanocage O32-15

EMDB-40209: 
Chlorophyll-binding region of de novo-designed nanocage O32-15

PDB-8glt: 
Backbone model of de novo-designed chlorophyll-binding nanocage O32-15

EMDB-41907: 
Computationally Designed, Expandable O4 Octahedral Handshake Nanocage
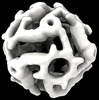
EMDB-42031: 
Computational Designed Nanocage O43_129_+8

EMDB-43318: 
Twistless helix 12 repeat ring design R12B

EMDB-37695: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH

EMDB-37696: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH

EMDB-37697: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH

EMDB-37698: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH

PDB-8woq: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at neutral pH

PDB-8wor: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at neutral pH

PDB-8wos: 
Cryo-EM structure of human SIDT1 protein with C1 symmetry at low pH

PDB-8wot: 
Cryo-EM structure of human SIDT1 protein with C2 symmetry at low pH

EMDB-29974: 
Cryo-EM structure of synthetic tetrameric building block sC4

EMDB-41364: 
CryoEM Structure of a Computationally Designed T3 Tetrahedral Nanocage

EMDB-42906: 
Computational Designed Nanocage O43_129

EMDB-42944: 
Computational Designed Nanocage O43_129_+4
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
