-Search query
-Search result
Showing 1 - 50 of 66 items for (author: hanc & p)

EMDB-39645: 
The structure of HKU1-B S protein with bsAb1

EMDB-39646: 
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein

PDB-8yww: 
The structure of HKU1-B S protein with bsAb1

PDB-8ywx: 
the complex structure of the H4B6 Fab with the RBD of Omicron BA.5 S protein

EMDB-40218: 
CryoEM structure of TnsC(1-503) bound to TnsD(1-318) from E.coli Tn7

EMDB-40221: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7

EMDB-40222: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7

EMDB-43138: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP

EMDB-43140: 
CyoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP

PDB-8glu: 
CryoEM structure of TnsC(1-503) bound to TnsD(1-318) from E.coli Tn7

PDB-8glw: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7

PDB-8glx: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7

PDB-8vcj: 
CryoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 7:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP

PDB-8vct: 
CyoEM structure of the TnsC(1-503)-TnsD(1-318)-DNA complex in a 6:2:1 stoichiometry from E. coli Tn7 bound to ATPgS and ADP

EMDB-43498: 
Mechanistic Insights Revealed by YbtPQ in the Occluded State

PDB-8vsi: 
Mechanistic Insights Revealed by YbtPQ in the Occluded State

EMDB-40856: 
Single particle reconstruction of the human LINE-1 ORF2p without substrate (apo)
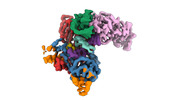
EMDB-40858: 
Structure of LINE-1 ORF2p with template:primer hybrid
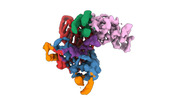
EMDB-40859: 
Structure of LINE-1 ORF2p with an oligo(A) template

PDB-8sxt: 
Structure of LINE-1 ORF2p with template:primer hybrid

PDB-8sxu: 
Structure of LINE-1 ORF2p with an oligo(A) template

EMDB-16120: 
Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to ATP and ADP

EMDB-16121: 
Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP

EMDB-16122: 
Cryo-EM structure of the wild-type solitary ECF module in MSP2N2 lipid nanodiscs in the ATPase open and nucleotide-free conformation

EMDB-16123: 
Cryo-EM structure of the wild-type solitary ECF module in DDM micelles in the ATPase open and nucleotide-free conformation

EMDB-16124: 
Cryo-EM structure of the mutant solitary ECF module 2EQ in MSP2N2 lipid nanodiscs in the ATPase closed and ATP-bound conformation

PDB-8bmp: 
Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to ATP and ADP

PDB-8bmq: 
Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to AMP-PNP

PDB-8bmr: 
Cryo-EM structure of the wild-type solitary ECF module in MSP2N2 lipid nanodiscs in the ATPase open and nucleotide-free conformation

PDB-8bms: 
Cryo-EM structure of the mutant solitary ECF module 2EQ in MSP2N2 lipid nanodiscs in the ATPase closed and ATP-bound conformation

EMDB-34259: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv282

EMDB-34260: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv282 focused on RBD_XGv282 sub-complex
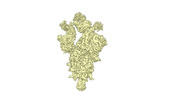
EMDB-34261: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv289

EMDB-34262: 
Cryo-EM map of Omicron BA.5 S protein in complex with XGv289 focused on RBD_XGv289 sub-complex
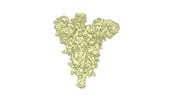
EMDB-34263: 
cryo-EM structure of Omicron BA.5 S protein in complex with S2L20

EMDB-34264: 
Cryo-EM map of Omicron BA.5 S protein in complex with S2L20 focused on NTD_S2L20 sub-complex

PDB-8gto: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv282

PDB-8gtp: 
cryo-EM structure of Omicron BA.5 S protein in complex with XGv289

PDB-8gtq: 
cryo-EM structure of Omicron BA.5 S protein in complex with S2L20

EMDB-27025: 
CRYO-EM STRUCTURE OF HUMAN 15-PGDH IN COMPLEX WITH SMALL MOLECULE SW222746

EMDB-29005: 
human 15-PGDH with NADH bound

PDB-8cwl: 
Cryo-EM structure of Human 15-PGDH in complex with small molecule SW222746

PDB-8fd8: 
human 15-PGDH with NADH bound

EMDB-27010: 
CRYO-EM STRUCTURE OF HUMAN 15-PGDH IN COMPLEX WITH SMALL MOLECULE SW209415

PDB-8cvn: 
Cryo-EM Structure of Human 15-PGDH in Complex with Small Molecule SW209415

EMDB-27243: 
Cryo-EM map of MORF-WHs in complex with 197bp nucleosome aided by scFv

EMDB-34046: 
Embigin facilitates monocarboxylate transporter 1 localization to plasma membrane and transition to a decoupling state

PDB-7yr5: 
Embigin facilitates monocarboxylate transporter 1 localization to plasma membrane and transition to a decoupling state

EMDB-26862: 
CryoEM structure of the TIR domain from AbTir in complex with 3AD

PDB-7uxu: 
CryoEM structure of the TIR domain from AbTir in complex with 3AD
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
