-Search query
-Search result
Showing all 25 items for (author: dixon & ne)

EMDB-44139: 
Structure of a complex between Pasteurella multocida surface lipoprotein, PmSLP-1, and bovine complement factor I
Method: single particle / : Nguyen QH, Norris MJ, Moraes TF

PDB-9b3h: 
Structure of a complex between Pasteurella multocida surface lipoprotein, PmSLP-1, and bovine complement factor I
Method: single particle / : Nguyen QH, Norris MJ, Moraes TF

EMDB-44146: 
Conformation 1 of the assembled dimer of PmSLP-1:FI complex
Method: single particle / : Nguyen QH, Norris MJ, Moraes TF

EMDB-44149: 
Conformation 2 of the assembled dimer of PmSLP-1:FI complex
Method: single particle / : Nguyen QH, Norris MJ, Moraes TF

EMDB-40079: 
E. coli clamp loader with closed clamp
Method: single particle / : Oakley AJ, Xu ZQ, Dixon NE

EMDB-40080: 
E. coli clamp loader with open clamp
Method: single particle / : Oakley AJ, Xu ZQ, Dixon NE

EMDB-40081: 
E. coli clamp loader with open clamp on primed template DNA (form 1)
Method: single particle / : Oakley AJ, Xu ZQ, Dixon NE

EMDB-40082: 
E. coli clamp loader with open clamp on primed template DNA (form 2)
Method: single particle / : Oakley AJ, Xu ZQ, Dixon NE

EMDB-40083: 
E. coli clamp loader with closed clamp on primed template DNA
Method: single particle / : Oakley AJ, Xu ZQ, Dixon NE

EMDB-40084: 
E. coli clamp loader on primed template DNA
Method: single particle / : Oakley AJ, Xu ZQ, Dixon NE

EMDB-25408: 
Mfd DNA complex
Method: single particle / : Oakley AJ, Xu ZQ

EMDB-25581: 
cryoEM reconstruction of the HIV gp140 in complex with the extracellular domains of CD4 and the adnectin domain of Combinectin. The gp140 and CD4 coordinates from entry 6EDU were rigid body fitted to the EM map along withe the crystal structure of CD4+adnectin
Method: single particle / : Concha NO, William SP

PDB-7t0o: 
cryoEM reconstruction of the HIV gp140 in complex with the extracellular domains of CD4 and the adnectin domain of Combinectin. The gp140 and CD4 coordinates from entry 6EDU were rigid body fitted to the EM map along withe the crystal structure of CD4+adnectin
Method: single particle / : Concha NO, William SP, Wenzel DL

EMDB-12876: 
Cryo-EM map of Scc2 bound to cohesin trimer
Method: single particle / : Gonzalez Llamazares A, Lee B, Lowe J

EMDB-12880: 
Scc2 bound to cohesin ATPase heads
Method: single particle / : Gonzalez Llamazares A, Lee B, Lowe J

EMDB-12887: 
Folded elbow of cohesin
Method: single particle / : Gonzalez Llamazares A, Lee BG, Collier J, Nasmyth KA, Lowe J

EMDB-12888: 
Pds5 bound to cohesin ATPase heads
Method: single particle / : Gonzalez Llamazares A, Lee B, Lowe J

EMDB-12889: 
Engaged ATPase heads of cohesin
Method: single particle / : Gonzalez Llamazares A, Lee B, Lowe J

PDB-7ogt: 
Folded elbow of cohesin
Method: single particle / : Lee BG, Gonzalez Llamazares A, Collier J, Patele NJ, Nasmyth KA, Lowe J

EMDB-4089: 
The human 26S Proteasome at 6.8 Ang.
Method: single particle / : Schweitzer A, Beck F

PDB-5ln3: 
The human 26S Proteasome at 6.8 Ang.
Method: single particle / : Schweitzer A, Beck F, Sakata E, Unverdorben P
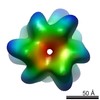
EMDB-1017: 
The DnaB.DnaC complex: a structure based on dimers assembled around an occluded channel.
Method: single particle / : Barcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher M, Carazo JM
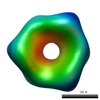
EMDB-1022: 
Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC.
Method: single particle / : San Martin C

EMDB-1023: 
Three-dimensional reconstructions from cryoelectron microscopy images reveal an intimate complex between helicase DnaB and its loading partner DnaC.
Method: single particle / : San Martin C
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
