-Search query
-Search result
Showing 1 - 50 of 95 items for (author: chen & jx)
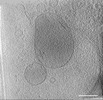
EMDB-39012: 
Representative tomogram of primary glioblastoma stem cell with circular inter-mitochondrial junctions.

EMDB-39015: 
Representative tomogram of microglia cell with nanotunnel-like structures resembling mitochondrial fission.
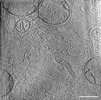
EMDB-39019: 
Representative tomogram of glioblastoma cell with nanotunnel-like structure and inter-mitochondrial junction.

EMDB-39021: 
Representative tomogram of normal human astrocyte with nanotunnel-like structure which is an extension of the mitochondrial outer membrane.

EMDB-39023: 
Representative tomogram of primary glioblastoma differentiated cell with parallel inter-mitochondrial junction.

EMDB-39024: 
Representative tomogram of primary glioblastoma stem cell with clustered mitochondria bearing various long-short axis ratios.

EMDB-38543: 
Cryo-EM map of the internal RuBisCOs in the alpha-carboxysome from Prochlorococcus MED4

EMDB-38544: 
Cryo-EM map of the intact shell of alpha-carboxysome from Prochlorococcus MED4

EMDB-37902: 
Cryo-EM structure of the alpha-carboxysome shell vertex from Prochlorococcus MED4

EMDB-37903: 
Cryo-EM map of the intact alpha-carboxysome from Prochlorococcus MED4

EMDB-33563: 
Structure of SUR2A in complex with Mg-ATP and repaglinide in the inward-facing conformation.

EMDB-33564: 
Structure of SUR2A in complex with Mg-ATP, Mg-ADP and repaglinide in the inward-facing conformation

EMDB-33565: 
Structure of SUR2B in complex with Mg-ATP and repaglinide in the inward-facing conformation

EMDB-33566: 
Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the inward-facing conformation

EMDB-33567: 
Structure of SUR2B in complex with Mg-ATP, Mg-ADP, and repaglinide in the partially occluded state
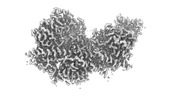
EMDB-15165: 
Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80

EMDB-34389: 
Structure of human phagocyte NADPH oxidase in the resting state

EMDB-34390: 
Consensus map of human phagocyte NADPH oxidase in the resting state

EMDB-15163: 
Cryo-EM reconstruction of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of INO80

EMDB-15164: 
Human Ino80 A-module + YY1
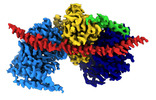
EMDB-15177: 
Cryo-EM reconstruction of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80

EMDB-15179: 
Cryo-EM reconstruction of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80
EMDB-15180: 
Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on curved DNA

EMDB-15184: 
Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 on straight DNA

EMDB-15186: 
Cryo-EM reconstruction of DNA bound Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80
EMDB-15187: 
A-module of Chaetomium thermophilum INO80 bound to curved DNA

EMDB-15188: 
Nucleosome bound Chaetomium thermophilum INO80 (ADP-BeF3 state) core complex with Arp5 grappler in parallel conformation

EMDB-15211: 
Ino80 core complex with A-module on nucleosome
EMDB-15647: 
Nucleosome-bound Ino80 ATPase
EMDB-15688: 
CryoEM structure of INO80 core nucleosome complex in closed grappler conformation (ADP-BeFx state)

EMDB-34620: 
Focus refined map of the constant regions of Fab heavy chain and light chain and TP1170 of human phagocyte NADPH oxidase in the resting state
EMDB-34621: 
Focus refined map of human phagocyte NADPH oxidase core in the resting state

EMDB-34622: 
Focus refined map of the DH domain of human phagocyte NADPH oxidase in the resting state

EMDB-32535: 
Structure of SUR1 in complex with mitiglinide

EMDB-33697: 
Cryo-EM structure of SARS-CoV-2 Omicron spike protein (S-6P-RRAR) in complex with human ACE2 ectodomain (one-RBD-up state)

EMDB-33698: 
Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (S-6P-RRAR)

EMDB-33690: 
Cryo-EM structure of apo SARS-CoV-2 Omicron spike protein (S-2P-GSAS)

EMDB-33699: 
Cryo-EM structure of hACE2-bound SARS-CoV-2 Omicron spike protein with L371S, P373S and F375S mutations (local refinement)

EMDB-33709: 
Cryo-EM structure of S309-RBD-RBD-S309 in the S309-bound Omicron spike protein (local refinement)

EMDB-32358: 
Structure of SARS-CoV-2 spike receptor-binding domain Y453F mutation complexed with American mink ACE2

EMDB-32379: 
Structure of SARS-CoV-2 spike receptor-binding domain F486L mutation complexed with American mink ACE2

EMDB-32310: 
The structure of KATP H175K mutant in pre-open state

EMDB-32311: 
The structure of KATP H175K mutant in closed state
EMDB-32024: 
Structure of SUR2B in complex with Mg-ATP/ADP

EMDB-32025: 
Structure of SUR2B in complex with MgATP/ADP and P1075

EMDB-32026: 
Structure of SUR2B in complex with Mg-ATP/ADP and levcromakalim

EMDB-32027: 
Structure of SUR2A in complex with Mg-ATP/ADP and P1075

EMDB-30903: 
Structure of TRPC3 at 2.7 angstrom in high calcium state

EMDB-30904: 
Structure of TRPC3 at 3.06 angstrom in low calcium state

EMDB-30906: 
Structure of TRPC3 gain of function mutation R803C at 3.2 angstrom in 1340nM free calcium state
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
