[English] 日本語
 Yorodumi
Yorodumi- EMDB-15186: Cryo-EM reconstruction of DNA bound Arp4-Ies4-N-actin-Arp8-Ino80H... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of DNA bound Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80 | ||||||||||||||||||
 Map data Map data | map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.5 Å | ||||||||||||||||||
 Authors Authors | Kunert F / Metzner FJ / Eustermann S / Jung J / Woike S / Schall K / Kostrewa D / Hopfner KP | ||||||||||||||||||
| Funding support | European Union,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural mechanism of extranucleosomal DNA readout by the INO80 complex. Authors: Franziska Kunert / Felix J Metzner / James Jung / Markus Höpfler / Stephan Woike / Kevin Schall / Dirk Kostrewa / Manuela Moldt / Jia-Xuan Chen / Susanne Bantele / Boris Pfander / Sebastian ...Authors: Franziska Kunert / Felix J Metzner / James Jung / Markus Höpfler / Stephan Woike / Kevin Schall / Dirk Kostrewa / Manuela Moldt / Jia-Xuan Chen / Susanne Bantele / Boris Pfander / Sebastian Eustermann / Karl-Peter Hopfner /  Abstract: The nucleosomal landscape of chromatin depends on the concerted action of chromatin remodelers. The INO80 remodeler specifically places nucleosomes at the boundary of gene regulatory elements, which ...The nucleosomal landscape of chromatin depends on the concerted action of chromatin remodelers. The INO80 remodeler specifically places nucleosomes at the boundary of gene regulatory elements, which is proposed to be the result of an ATP-dependent nucleosome sliding activity that is regulated by extranucleosomal DNA features. Here, we use cryo-electron microscopy and functional assays to reveal how INO80 binds and is regulated by extranucleosomal DNA. Structures of the regulatory A-module bound to DNA clarify the mechanism of linker DNA binding. The A-module is connected to the motor unit via an HSA/post-HSA lever element to chemomechanically couple the motor and linker DNA sensing. Two notable sites of curved DNA recognition by coordinated action of the four actin/actin-related proteins and the motor suggest how sliding by INO80 can be regulated by extranucleosomal DNA features. Last, the structures clarify the recruitment of YY1/Ies4 subunits and reveal deep architectural similarities between the regulatory modules of INO80 and SWI/SNF complexes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15186.map.gz emd_15186.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15186-v30.xml emd-15186-v30.xml emd-15186.xml emd-15186.xml | 19.8 KB 19.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15186.png emd_15186.png | 59.8 KB | ||
| Others |  emd_15186_half_map_1.map.gz emd_15186_half_map_1.map.gz emd_15186_half_map_2.map.gz emd_15186_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15186 http://ftp.pdbj.org/pub/emdb/structures/EMD-15186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15186 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15186 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_15186.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15186.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
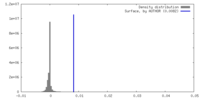
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_15186_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
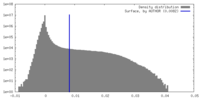
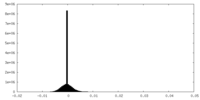
| Density Histograms |
-Half map: half map B
| File | emd_15186_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
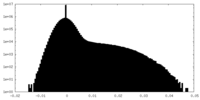
| Density Histograms |
- Sample components
Sample components
-Entire : INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) bound to DNA
| Entire | Name: INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) bound to DNA |
|---|---|
| Components |
|
-Supramolecule #1: INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) bound to DNA
| Supramolecule | Name: INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) bound to DNA type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Ino80
| Macromolecule | Name: Ino80 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSLAVLLNKE DKDISDFSKT TAGKSAKKNS RERVADVAPT RVLDKKQAYL SQLNSEFNRI KRRDSIEQLY QDWKFINLQE FELISEWNQQ SKDWQFDNTN DSQDLHFKKL YRDMSMINKE WAEYQSFKNA NLSDIINEKD ADEDEEDDED ELEDGEEDME EDEASTGRHT ...String: MSLAVLLNKE DKDISDFSKT TAGKSAKKNS RERVADVAPT RVLDKKQAYL SQLNSEFNRI KRRDSIEQLY QDWKFINLQE FELISEWNQQ SKDWQFDNTN DSQDLHFKKL YRDMSMINKE WAEYQSFKNA NLSDIINEKD ADEDEEDDED ELEDGEEDME EDEASTGRHT NGKSMRGNGI QKSRKKDAAA AAAIGKAIKD DQTHADTVVT VNGDENEDGN NGEDEDNDND NENNNDNDND NENENDNDSD NDDEEENGEE DEEEEEIEDL DEEDFAAFEE QDDNDDEDFN PDVEKRRKRS SSSSSSTKLS MNSLSLITSK KINKNITINS DRPKIVRELI KMCNKNKHQK IKKRRFTNCI VTDYNPIDSK LNIKITLKQY HVKRLKKLIN DAKREREREE ALKNNVGLDG NDLDNDEDGS ESHKRRKLNN NTANGADDAN KRKFNTRHGL PTYGMKMNAK EARAIQRHYD NTYTTIWKDM ARKDSTKMSR LVQQIQSIRS TNFRKTSSLC AREAKKWQSK NFKQIKDFQT RARRGIREMS NFWKKNEREE RDLKKKIEKE AMEQAKKEEE EKESKRQAKK LNFLLTQTEL YSHFIGRKDY KDDDDKGTDY KDDDDK |
-Macromolecule #2: Arp8
| Macromolecule | Name: Arp8 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSQEEAESSI IYEEPIDIPL EDDDDEDELE EENSVPLSSQ ADQENAENES DDSVDNVVGS ETPRSVTGLS VDPRDVADEE DEDEEGEDED EDEDDNDVDN EDENDNDNAN ENENELGSSR DKRAPPAVQT SKRYKKYPKL DPAKAPPGKK VPLHLLEKRR LGRIKAAEEF ...String: MSQEEAESSI IYEEPIDIPL EDDDDEDELE EENSVPLSSQ ADQENAENES DDSVDNVVGS ETPRSVTGLS VDPRDVADEE DEDEEGEDED EDEDDNDVDN EDENDNDNAN ENENELGSSR DKRAPPAVQT SKRYKKYPKL DPAKAPPGKK VPLHLLEKRR LGRIKAAEEF AKTLKKIGIE KVETTTLPAT GLFQPLMLIN QKNYSSDYLK KDDQIFALRD RKFLRNNNTS QISSTNTPDV IDLKSLPHSE ASAAPLNDEI DLNDPTATIV IHPGSNSIKI GFPKDDHPVV VPNCVAVPKK WLDLENSEHV ENVCLQREQS EEFNNIKSEM EKNFRERMRY YKRKVPGNAH EQVVSFNENS KPEIISEKND PSPIEWIFDD SKLYYGSDAL RCVDEKFVIR KPFRGGSFNV KSPYYKSLAE LISDVTKLLE HALNSETLNV KPTKFNQYKV VLVIPDIFKK SHVETFIRVL LTELQFQAVA IIQESLATCY GAGISTSTCV VNIGAAETRI ACVDEGTVLE HSAITLDYGG DDITRLFALF LLQSDFPLQD WKIDSKHGWL LAERLKKNFT TFQDADVAVQ LYNFMNRSPN QPTEKYEFKL FDEVMLAPLA LFFPQIFKLI RTSSHKNSSL EFQLPESRDL FTNELNDWNS LSQFESKEGN LYCDLNDDLK ILNRILDAHN IIDQLQDKPE NYGNTLKENF APLEKAIVQS IANASITADV TRMNSFYSNI LIVGGSSKIP ALDFILTDRI NIWRPSLLSS ASFPQFYKKL TKEIKDLEGH YVNAPDKTED ENKQILQAQI KEKIVEELEE QHQNIEHQNG NEHIFPVSII PPPRDMNPAL IIWKGASVLA QIKLVEELFI TNSDWDVHGS RILQYKCIFT Y |
-Macromolecule #3: Actin
| Macromolecule | Name: Actin / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MDSEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGIMVGMGQK DSYVGDEAQS KRGILTLRYP IEHGIVTNWD DMEKIWHHTF YNELRVAPEE HPVLLTEAPM NPKSNREKMT QIMFETFNVP AFYVSIQAVL SLYSSGRTTG IVLDSGDGVT HVVPIYAGFS ...String: MDSEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH QGIMVGMGQK DSYVGDEAQS KRGILTLRYP IEHGIVTNWD DMEKIWHHTF YNELRVAPEE HPVLLTEAPM NPKSNREKMT QIMFETFNVP AFYVSIQAVL SLYSSGRTTG IVLDSGDGVT HVVPIYAGFS LPHAILRIDL AGRDLTDYLM KILSERGYSF STTAEREIVR DIKEKLCYVA LDFEQEMQTA AQSSSIEKSY ELPDGQVITI GNERFRAPEA LFHPSVLGLE SAGIDQTTYN SIMKCDVDVR KELYGNIVMS GGTTMFPGIA ERMQKEITAL APSSMKVKII APPERKYSVW IGGSILASLT TFQQMWISKQ EYDESGPSIV HHKCF |
-Macromolecule #4: Arp4
| Macromolecule | Name: Arp4 / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSNAALQVYG GDEVSAVVID PGSYTTNIGY SGSDFPQSIL PSVYGKYTAD EGNKKIFSEQ SIGIPRKDYE LKPIIENGLV IDWDTAQEQW QWALQNELYL NSNSGIPALL TEPVWNSTEN RKKSLEVLLE GMQFEACYLA PTSTCVSFAA GRPNCLVVDI GHDTCSVSPI ...String: MSNAALQVYG GDEVSAVVID PGSYTTNIGY SGSDFPQSIL PSVYGKYTAD EGNKKIFSEQ SIGIPRKDYE LKPIIENGLV IDWDTAQEQW QWALQNELYL NSNSGIPALL TEPVWNSTEN RKKSLEVLLE GMQFEACYLA PTSTCVSFAA GRPNCLVVDI GHDTCSVSPI VDGMTLSKST RRNFIAGKFI NHLIKKALEP KEIIPLFAIK QRKPEFIKKT FDYEVDKSLY DYANNRGFFQ ECKETLCHIC PTKTLEETKT ELSSTAKRSI ESPWNEEIVF DNETRYGFAE ELFLPKEDDI PANWPRSNSG VVKTWRNDYV PLKRTKPSGV NKSDKKVTPT EEKEQEAVSK STSPAANSAD TPNETGKRPL EEEKPPKENN ELIGLADLVY SSIMSSDVDL RATLAHNVVL TGGTSSIPGL SDRLMTELNK ILPSLKFRIL TTGHTIERQY QSWLGGSILT SLGTFHQLWV GKKEYEEVGV ERLLNDRFR |
-Macromolecule #5: Ies4
| Macromolecule | Name: Ies4 / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: MSQESSVLSE SQEQLANNPK IEDTSPPSAN SRDNSKPVLP WDYKNKAIEI KSFSGYKVNF TGWIRRDVRE ERQRGSEFTA SDVKGSDDKA TRKKEPADED PEVKQLEKEG EDGLDS |
-Macromolecule #6: DNA
| Macromolecule | Name: DNA / type: dna / ID: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  |
| Sequence | String: CTTACCCTGC GTGCCCGCGC ACCCTGGCGA CTTCGCCTCG TTTTGGCGAT TTTCTTAG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 44.68 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 69226 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)