[English] 日本語
 Yorodumi
Yorodumi- EMDB-15165: Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of Chaetomium thermophilum INO80 | ||||||||||||||||||
 Map data Map data | main map | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | chromatin remodeler / INO80 / Actin-related protein / DNA binding protein | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationIno80 complex / chromatin organization / chromatin remodeling / DNA repair / nucleus Similarity search - Function | ||||||||||||||||||
| Biological species |  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | ||||||||||||||||||
 Authors Authors | Kunert F / Metzner FJ / Eustermann S / Jung J / Woike S / Schall K / Kostrewa D / Hopfner KP | ||||||||||||||||||
| Funding support | European Union,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2022 Journal: Sci Adv / Year: 2022Title: Structural mechanism of extranucleosomal DNA readout by the INO80 complex. Authors: Franziska Kunert / Felix J Metzner / James Jung / Markus Höpfler / Stephan Woike / Kevin Schall / Dirk Kostrewa / Manuela Moldt / Jia-Xuan Chen / Susanne Bantele / Boris Pfander / Sebastian ...Authors: Franziska Kunert / Felix J Metzner / James Jung / Markus Höpfler / Stephan Woike / Kevin Schall / Dirk Kostrewa / Manuela Moldt / Jia-Xuan Chen / Susanne Bantele / Boris Pfander / Sebastian Eustermann / Karl-Peter Hopfner /  Abstract: The nucleosomal landscape of chromatin depends on the concerted action of chromatin remodelers. The INO80 remodeler specifically places nucleosomes at the boundary of gene regulatory elements, which ...The nucleosomal landscape of chromatin depends on the concerted action of chromatin remodelers. The INO80 remodeler specifically places nucleosomes at the boundary of gene regulatory elements, which is proposed to be the result of an ATP-dependent nucleosome sliding activity that is regulated by extranucleosomal DNA features. Here, we use cryo-electron microscopy and functional assays to reveal how INO80 binds and is regulated by extranucleosomal DNA. Structures of the regulatory A-module bound to DNA clarify the mechanism of linker DNA binding. The A-module is connected to the motor unit via an HSA/post-HSA lever element to chemomechanically couple the motor and linker DNA sensing. Two notable sites of curved DNA recognition by coordinated action of the four actin/actin-related proteins and the motor suggest how sliding by INO80 can be regulated by extranucleosomal DNA features. Last, the structures clarify the recruitment of YY1/Ies4 subunits and reveal deep architectural similarities between the regulatory modules of INO80 and SWI/SNF complexes. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15165.map.gz emd_15165.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15165-v30.xml emd-15165-v30.xml emd-15165.xml emd-15165.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15165.png emd_15165.png | 60.3 KB | ||
| Filedesc metadata |  emd-15165.cif.gz emd-15165.cif.gz | 7.1 KB | ||
| Others |  emd_15165_half_map_1.map.gz emd_15165_half_map_1.map.gz emd_15165_half_map_2.map.gz emd_15165_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15165 http://ftp.pdbj.org/pub/emdb/structures/EMD-15165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15165 | HTTPS FTP |
-Related structure data
| Related structure data |  8a5dMC  8a5aC  8a5oC  8a5pC  8a5qC  8atfC  8av6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15165.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15165.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.059 Å | ||||||||||||||||||||||||||||||||||||
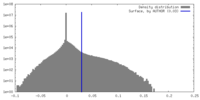
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_15165_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
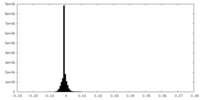
| Density Histograms |
-Half map: half map B
| File | emd_15165_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
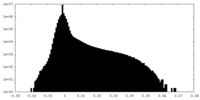
| Density Histograms |
- Sample components
Sample components
-Entire : Chaetomium thermophilum INO80 A-Module (Arp4-Ies4-N-actin-Arp8-In...
| Entire | Name: Chaetomium thermophilum INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) |
|---|---|
| Components |
|
-Supramolecule #1: Chaetomium thermophilum INO80 A-Module (Arp4-Ies4-N-actin-Arp8-In...
| Supramolecule | Name: Chaetomium thermophilum INO80 A-Module (Arp4-Ies4-N-actin-Arp8-Ino80HSA) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) Thermochaetoides thermophila (fungus) |
-Macromolecule #1: Ino80 ATPase
| Macromolecule | Name: Ino80 ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 37.445441 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: RKRRREKSMA ATLEQQAAAL QKASQAEDEA ERLKYIREAE RANKKVQQTK YILQKGIKGP ARNLPPIEPN LEGGTMATFS AENMEPGKV KGKGRGGRLK KSKEQKQAEK EQAELAQAAI DAGEEPPSKE ETKIRIKLSK AKASAKEEAE KDKENKEPKQ P KEPEKPVE ...String: RKRRREKSMA ATLEQQAAAL QKASQAEDEA ERLKYIREAE RANKKVQQTK YILQKGIKGP ARNLPPIEPN LEGGTMATFS AENMEPGKV KGKGRGGRLK KSKEQKQAEK EQAELAQAAI DAGEEPPSKE ETKIRIKLSK AKASAKEEAE KDKENKEPKQ P KEPEKPVE EPKDPLELKF QSKGYNQIYD QIWRDLARKD VSKVFRLATD SYATKASNLK KTAILASKEA KRWQLRTNKG TK DLQARAK RVMRDMMGFW KRNEREERDL RKAAERLELE NARKEEADRE AARQRRKLNF LISQTELYSD YKDDDDKGTD YKD DDDK |
-Macromolecule #2: Actin-related protein 8
| Macromolecule | Name: Actin-related protein 8 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 84.139266 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVGKVSERVL AREGLERTDN GMRQTSWPEV QPINQKNYYT DYMKRDDQIL SLRLQNEANR ERLVKTAQDR DRALNTNTNG EVPLPLDEL QEDAPPAPSA AMDPSKIIVI HPGSQNLRIG FASDALPKTI PMVLGTQYSQ TESEMHEALP RRQFEGRTME Q QYGEEWVK ...String: MVGKVSERVL AREGLERTDN GMRQTSWPEV QPINQKNYYT DYMKRDDQIL SLRLQNEANR ERLVKTAQDR DRALNTNTNG EVPLPLDEL QEDAPPAPSA AMDPSKIIVI HPGSQNLRIG FASDALPKTI PMVLGTQYSQ TESEMHEALP RRQFEGRTME Q QYGEEWVK KYQKMCSDLK VTMRANKLKV LPNSKDLVVN FNRRTEPEII SQHNDPLQVE WTNVNKAPED GTTQKVFIGQ QA LRIAEDS SPKYKLWWPI QHGWLNEDDY PTRAHLFDDL EMLLDRALRR ELGLTKKADW KQYSCVIVIP DLYDKRYVEL LLH LCIEFF DLSRVAFIQE SMAATFGAGY TQACVVDVGA QKTSISCVED GLVIEDSRVN LKYGGYDVTE TFIKMMLYDN FPYQ DINLR RRHDFLLAEE LKMKYCTLSQ ANISVQAFDF HLRAPNQPTR KYQFKFYDEV ILAPMGFYDP SIFDNSTKLR GRRKL IDRS YNAYDVDMPD DPTSSAQLAI LAMVQPSLAA PTATASFNGD PTATPSKERT QPFNFLSRPD ATGTPGTSKA PSPAPD GAS TPVPGPYIFG ASSREANGGS PAPSGRNGGT PAPANGTTVL PATSFDNPTS QQPQQRSAKE IAAERDAVLP IAPLDVA IL TSIQHAAKGD EKKVRELVGS IMVVGGGAKI PHFAPFLEEK LKIRRPDLAD RILVSRSARE MDEQVVVWKG ASVFAKLS T NDSWVTNYEY KMLGSRCIYM KLLWHY UniProtKB: Putative actin-related protein |
-Macromolecule #3: Actin
| Macromolecule | Name: Actin / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 41.735512 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEEEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH HGIMIGMGQK DSYVGDEAQS KRGILTLRYP IEHGVVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP INPKSNREKM TQIVFETFNA PAFYVSIQAV LSLYASGRTT GIVLDSGDGV T HVVPIYEG ...String: MEEEVAALVI DNGSGMCKAG FAGDDAPRAV FPSIVGRPRH HGIMIGMGQK DSYVGDEAQS KRGILTLRYP IEHGVVTNWD DMEKIWHHT FYNELRVAPE EHPVLLTEAP INPKSNREKM TQIVFETFNA PAFYVSIQAV LSLYASGRTT GIVLDSGDGV T HVVPIYEG FSLPHAIARL DMAGRDLTDY LMKILAERGY TFSTTAEREI VRDIKEKLCY VALDFEQEIQ TAAQSSHLEK SY ELPDGQV ITIGNERFRA PEALFQPSVL GLESGGIHVT TFNSIMKCDV DVRKDLYGNI VMSGGTTMYP GLSDRMQKEI TAL APSSMK VKIIAPPERK YSVWIGGSIL ASLSTFQQMW ISKQEYDESG PSIVHRKCF UniProtKB: Actin |
-Macromolecule #4: Actin related protein 4 (Arp4)
| Macromolecule | Name: Actin related protein 4 (Arp4) / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 51.761199 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MSQQPLSSSV QPADIYGGDE VSALVLDPGY CNTRAGFAGE EMPKQVIPSF YGHVDGRDLF GDEVIVPRAG FEVRNYMNRD SMVEDWDAA TRVWEYLLVQ RLQPPRPTPA SKNGLNVSDD GDVQMEDSAP NEDALDALEK PLTENPLLMT EAPWNTPKAR E KAIEVVME ...String: MSQQPLSSSV QPADIYGGDE VSALVLDPGY CNTRAGFAGE EMPKQVIPSF YGHVDGRDLF GDEVIVPRAG FEVRNYMNRD SMVEDWDAA TRVWEYLLVQ RLQPPRPTPA SKNGLNVSDD GDVQMEDSAP NEDALDALEK PLTENPLLMT EAPWNTPKAR E KAIEVVME NWGTPAFWLS RTPVLAAFAA GKATALVIDV GGANTSVTAI HDGMVLKRSI QRSPAAGVWL SGQIRSMWKS QD PPVNVVP TFMVENKKPV EAGAPPDCRL RNFGFPIHDS FRAFEEERVL TEFKESVVEV WRGPGKYLNP GNEDFAKTQP GRV FEFPDG SNQMWREQRY RVAEGMWDET AAYPSLNPDE AAITKAQTIP ALIKAALDGV DVDLRPNLLG NVVVTGSTSL LNGF NDRLN HELTNMYPGL KIKLHAAGLT SERRFGAWIG GSILASLGTF HQMWISRKEY EENGAGIVEK RCK UniProtKB: Actin-related protein 4 |
-Macromolecule #5: Ies4
| Macromolecule | Name: Ies4 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 Thermochaetoides thermophila (fungus) / Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 29.015457 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAPSSKNATP SNKDRRKSGI AAAGNGNAAA SSKIVTLKVT PERLRAIFDP DILMEDAPAS KASPAAANSN TENATESNPN SPSASTPAG QSVMGPPAEG PKKKGVKRGA AALGPNGEPK PRGKPGPKKK QRLEDGTIVE EPSRKLGPKA NQGAINAGLR A LDRSGKPC ...String: MAPSSKNATP SNKDRRKSGI AAAGNGNAAA SSKIVTLKVT PERLRAIFDP DILMEDAPAS KASPAAANSN TENATESNPN SPSASTPAG QSVMGPPAEG PKKKGVKRGA AALGPNGEPK PRGKPGPKKK QRLEDGTIVE EPSRKLGPKA NQGAINAGLR A LDRSGKPC RKWTRGTFQM KSFTGVVWEI PRWTAPPRPK PETAAPDDAA ATVAVNGNGA TPASTAADNN SNKENNNPSI AA AKVEDAS SQTAVKSEMS TGSGDIEMQS VAPSIAPSPA PVPIAVA UniProtKB: INO80 complex subunit 4 |
-Macromolecule #6: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 6 / Number of copies: 3 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #7: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 7 / Number of copies: 3 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 44.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.9 µm / Nominal defocus min: 1.1 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 343000 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)