-Search query
-Search result
Showing 1 - 50 of 1,408 items for (author: chang & sh)

EMDB-50621: 
Structure of heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 2)

EMDB-50628: 
Structure of heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 1)

PDB-9fof: 
Structure of heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 2)

PDB-9for: 
Structure of heteromeric amyloid filament of TDP-43 and AXNA11 from FTLD-TDP Type C (variant 1)

EMDB-37467: 
SARS-CoV-2 Omicron BQ.1.1 RBD complexed with human ACE2

EMDB-37468: 
SARS-CoV-2 Omicron BQ.1 RBD complexed with human ACE2

EMDB-37469: 
SARS-CoV-2 Omicron XBB RBD complexed with human ACE2

EMDB-37470: 
SARS-CoV-2 Omicron BF.7 RBD complexed with human ACE2

EMDB-37471: 
SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304

PDB-8wdy: 
SARS-CoV-2 Omicron BQ.1.1 RBD complexed with human ACE2

PDB-8wdz: 
SARS-CoV-2 Omicron BQ.1 RBD complexed with human ACE2

PDB-8we0: 
SARS-CoV-2 Omicron XBB RBD complexed with human ACE2

PDB-8we1: 
SARS-CoV-2 Omicron BF.7 RBD complexed with human ACE2

PDB-8we4: 
SARS-CoV-2 Omicron XBB.1.5 RBD complexed with human ACE2 and S304

EMDB-18180: 
cryoEM structure of SARS-CoV2 Spike trimer in complex with Fab23

PDB-8q5y: 
cryoEM structure of SARS-CoV2 Spike trimer in complex with Fab23

EMDB-37593: 
Vibrio vulnificus MARTX effector duet (RDTND-RID) complexed with human Rac1 Q61L and calmodulin
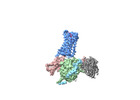
EMDB-39901: 
cryo-EM structure of the octreotide-bound SSTR5-Gi complex
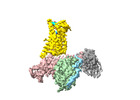
EMDB-39931: 
Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex

PDB-8zbe: 
cryo-EM structure of the octreotide-bound SSTR5-Gi complex

PDB-8zcj: 
Cryo-EM structure of the pasireotide-bound SSTR5-Gi complex

EMDB-38460: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state

EMDB-38463: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state

EMDB-38476: 
Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state

EMDB-38488: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state

EMDB-38495: 
SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein)

EMDB-38496: 
SARS-CoV-2 Omicron HV.1 RBD in complex with human ACE2 (local refinement from the spike protein)

EMDB-38498: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2

EMDB-38502: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2

EMDB-38505: 
Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2

EMDB-38826: 
Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2

EMDB-38827: 
Cryo-EM structure of SARS-CoV-2 Omicron JN.1 RBD in complex with human ACE2 (local refinement from the spike protein)

EMDB-38937: 
Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein

EMDB-38983: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 and S309 Fab

PDB-8xlv: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state

PDB-8xm5: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state

PDB-8xmg: 
Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state

PDB-8xmt: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state

PDB-8xn2: 
SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein)

PDB-8xn3: 
SARS-CoV-2 Omicron HV.1 RBD in complex with human ACE2 (local refinement from the spike protein)

PDB-8xn5: 
Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2

PDB-8xnf: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2

PDB-8xnk: 
Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2

PDB-8y16: 
Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2

PDB-8y18: 
Cryo-EM structure of SARS-CoV-2 Omicron JN.1 RBD in complex with human ACE2 (local refinement from the spike protein)

PDB-8y5j: 
Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein

PDB-8y6a: 
Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 and S309 Fab

EMDB-37210: 
Prefusion RSV F Bound to Lonafarnib and D25 Fab

PDB-8kg5: 
Prefusion RSV F Bound to Lonafarnib and D25 Fab

EMDB-38216: 
Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with antibody O5C2
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
