-Search query
-Search result
Showing 1 - 50 of 301 items for (author: beat & f)
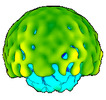
EMDB-50358: 
In vitro-induced genome-releasing intermediate of Rhodobacter microvirus Ebor computed with C5 symmetry

EMDB-50356: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-50357: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry
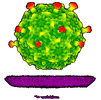
EMDB-50359: 
Rhodobacter microvirus Ebor attached to B10 host cell reconstructed by single particle analysis with applied C5 symmetry
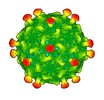
EMDB-50360: 
Rhodobacter microvirus Ebor attached to the outer membrane vesicle

EMDB-50361: 
Rhodobacter microvirus Ebor attached to the host cell reconstructed by subtomogram averaging

PDB-9ffg: 
Empty capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

PDB-9ffh: 
Native capsid of Rhodobacter microvirus Ebor computed with I4 symmetry

EMDB-17863: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod

EMDB-17878: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod

PDB-8psv: 
2.7 A cryo-EM structure of in vitro assembled type 1 pilus rod

PDB-8ptu: 
2.5 A cryo-EM structure of the in vitro FimD-catalyzed assembly of type 1 pilus rod

EMDB-43658: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs

EMDB-43659: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement

EMDB-43660: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement

PDB-8vye: 
SARS-CoV-2 S (C.37 Lambda variant) plus S309, S2L20, and S2X303 Fabs

PDB-8vyf: 
SARS-CoV-2 S NTD (C.37 Lambda variant) plus S2L20 and S2X303 Fabs, local refinement

PDB-8vyg: 
SARS-CoV-2 S RBD (C.37 Lambda variant) plus S309 Fab, local refinement
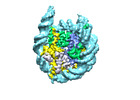
EMDB-18699: 
Human H3 nucleosome assembled on alpha-satellite DNA (Class1: most wrapped DNA)
EMDB-18714: 
Human H3 nucleosome assembled on alpha-satellite DNA (most unwrapped)
EMDB-18739: 
Human CENP-A nucleosome assembled on alpha-satellite DNA (most wrapped DNA)
EMDB-18740: 
Human CENP-A nucleosome assembled on alpha-satellite DNA (partially unwrapped)

EMDB-18745: 
Human Cenp-A nucleosome assembled on alpha-satellite DNA (most unwrapped DNA)
EMDB-18753: 
Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (most wrapped DNA)
EMDB-18763: 
Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA)
EMDB-18768: 
Human CENP-A nucleosome assembled on alpha-satellite DNA (most unwrapped DNA)
EMDB-18775: 
Human CENP-A nucleosome assembled on 601 DNA with CENP-B box
EMDB-18776: 
Human CENP-A nucleosome assembled on 601 DNA with CENP-B box in complex with CENP-B

EMDB-16512: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-16522: 
Structure of ADDoCoV-ADAH11

PDB-8c9n: 
MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle

EMDB-14778: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation

EMDB-14801: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry

EMDB-14852: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry

EMDB-14960: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation

PDB-7zla: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the half-closed conformation

PDB-7zn5: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C2 symmetry.

PDB-7zpa: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the closed conformation, C1 symmetry

PDB-7zth: 
Cryo-EM structure of holo-PdxR from Bacillus clausii bound to its target DNA in the open conformation

EMDB-15118: 
13pf undecorated microtubule from recombinant human tubulin (alpha1B, beta3) lacking the C-terminal tail

EMDB-15119: 
13pf undecorated microtubule from recombinant human tubulin (alpha1B, beta3) with spliced unmodified C-terminal tail on alpha1B.

EMDB-15120: 
13pf undecorated microtubule from recombinant human tubulin (alpha1B, beta3) with spliced C-terminal tail containing 10E branch on alpha1B.

EMDB-17154: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (consensus and constituent map 1)

EMDB-17155: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 1)

EMDB-17156: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL-6.2 (DNA conformation 2)

EMDB-17157: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (composite map)

EMDB-17158: 
Cryo-EM structure of CLOCK-BMAL1 bound to a nucleosomal E-box at position SHL+5.8 (constituent map 2 from additional focus classification on PAS domains)

EMDB-17159: 
Cryo-EM map of MYC-MAX-OCT4-LIN28 complex

EMDB-17160: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 2, additional 3D classification and flexible refinement)

EMDB-17161: 
Cryo-EM structure of CLOCK-BMAL1 bound to the native Por enhancer nucleosome (map 1)
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
