[English] 日本語
 Yorodumi
Yorodumi- EMDB-18763: Human CENP-A nucleosome assembled on alpha-satellite DNA in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
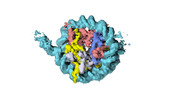
| Title | Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA) | |||||||||
 Map data Map data | Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nucleosome / centromere / centromeric / CENP-A / Histones / Human / DNA / DNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
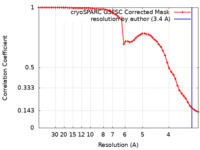
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Ali-Ahmad A / Sekulic N | |||||||||
| Funding support |  Norway, 2 items Norway, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: CENP-A and CENP-B collaborate to create an open centromeric chromatin state. Authors: Harsh Nagpal / Ahmad Ali-Ahmad / Yasuhiro Hirano / Wei Cai / Mario Halic / Tatsuo Fukagawa / Nikolina Sekulić / Beat Fierz /     Abstract: Centromeres are epigenetically defined via the presence of the histone H3 variant CENP-A. Contacting CENP-A nucleosomes, the constitutive centromere associated network (CCAN) and the kinetochore ...Centromeres are epigenetically defined via the presence of the histone H3 variant CENP-A. Contacting CENP-A nucleosomes, the constitutive centromere associated network (CCAN) and the kinetochore assemble, connecting the centromere to spindle microtubules during cell division. The DNA-binding centromeric protein CENP-B is involved in maintaining centromere stability and, together with CENP-A, shapes the centromeric chromatin state. The nanoscale organization of centromeric chromatin is not well understood. Here, we use single-molecule fluorescence and cryoelectron microscopy (cryoEM) to show that CENP-A incorporation establishes a dynamic and open chromatin state. The increased dynamics of CENP-A chromatin create an opening for CENP-B DNA access. In turn, bound CENP-B further opens the chromatin fiber structure and induces nucleosomal DNA unwrapping. Finally, removal of CENP-A increases CENP-B mobility in cells. Together, our studies show that the two centromere-specific proteins collaborate to reshape chromatin structure, enabling the binding of centromeric factors and establishing a centromeric chromatin state. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_18763.map.gz emd_18763.map.gz | 8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-18763-v30.xml emd-18763-v30.xml emd-18763.xml emd-18763.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_18763_fsc.xml emd_18763_fsc.xml | 5.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_18763.png emd_18763.png | 45.2 KB | ||
| Masks |  emd_18763_msk_1.map emd_18763_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-18763.cif.gz emd-18763.cif.gz | 5.3 KB | ||
| Others |  emd_18763_half_map_1.map.gz emd_18763_half_map_1.map.gz emd_18763_half_map_2.map.gz emd_18763_half_map_2.map.gz | 14.5 MB 14.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-18763 http://ftp.pdbj.org/pub/emdb/structures/EMD-18763 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18763 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-18763 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_18763.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_18763.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.584 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_18763_msk_1.map emd_18763_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
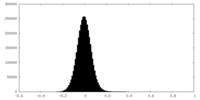
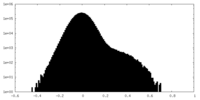
| Density Histograms |
-Half map: #2
| File | emd_18763_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_18763_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human CENP-A nucleosome assembled on alpha-satellite DNA in compl...
| Entire | Name: Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA) |
|---|---|
| Components |
|
-Supramolecule #1: Human CENP-A nucleosome assembled on alpha-satellite DNA in compl...
| Supramolecule | Name: Human CENP-A nucleosome assembled on alpha-satellite DNA in complex with CENP-B (partially unwrapped DNA) type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Human Histone CENP-A
| Macromolecule | Name: Human Histone CENP-A / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MARTKQTARK STGGKAPRKQ LATKAARKSA PATGGVKKPH RYRPGTVALR EIRKLQKSTH LLIRKLPFSR LAREICVKFT RGVDFNWQAQ ALLALQEAAE AFLVHLFEDA YLLTLHAGRV TLFPKDVQLA RRIRGLEEGL G |
-Macromolecule #2: Human histone H4
| Macromolecule | Name: Human histone H4 / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSGRGKGGKG LGKGGAKRHR KVLRDNIQGI TKPAIRRLAR RGGVKRISGL IYEETRGVLK VFLENVIRDA VTYTEHAKRK TVTAMDVVYA LKRQGRTLYG FGG |
-Macromolecule #3: Human histone H2A
| Macromolecule | Name: Human histone H2A / type: protein_or_peptide / ID: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPLGMSGRGK QGGKARAKAK SRSSRAGLQF PVGRVHRLLR KGNYAERVGA GAPVYMAAVL EYLTAEILEL AGNAARDNKK TRIIPRHLQL AIRNDEELNK LLGKVTIAQG GVLPNIQAVL LPKKTESHHK AKGK |
-Macromolecule #4: Human histone H2B
| Macromolecule | Name: Human histone H2B / type: protein_or_peptide / ID: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MPEPAKSAPA PKKGSKKAVT KAQKKDGKKR KRSRKESYSV YVYKVLKQVH PDTGISSKAM GIMNSFVNDI FERIAGEASR LAHYNKRSTI TSREIQTAVR LLLPGELAKH AVSEGTKAVT KYTSSK |
-Macromolecule #5: Human CENP-B
| Macromolecule | Name: Human CENP-B / type: protein_or_peptide / ID: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DSLEFIASKL AGPKRRQLTF REKSRIIQEV EENPDLRKGE IARRFNIPPS TLSTILKNKR AILASERKYG VASTCRKTNK LSPYDKLEGL LIAWFQQIRA AGLPVKGIIL KEKALRIAEE LGMDDFTASN GWLDRFRRRH GVVSCSGVAR ARARNAAPRT PAAPASPAAV ...String: DSLEFIASKL AGPKRRQLTF REKSRIIQEV EENPDLRKGE IARRFNIPPS TLSTILKNKR AILASERKYG VASTCRKTNK LSPYDKLEGL LIAWFQQIRA AGLPVKGIIL KEKALRIAEE LGMDDFTASN GWLDRFRRRH GVVSCSGVAR ARARNAAPRT PAAPASPAAV PSEGSGGSTT GWRAREEQPP SVAEGYASQD VFSATETSLW YDFLPDQAAG LCGGDGRPRQ ATQRLSVLLC ANADGSEKLP PLVAGKSAKP RAGQAGLPCD YTANSKGGVT TQALAKYLKA LDTRMAAESR RVLLLAGRLA AQSLDTSGLR HVQLAFFPPG TVHPLERGVV QQVKGHYRQA MLLKAMAALE GQDPSGLQLG LTEALHFVAA AWQAVEPSDI AACFREAGFG GGPNATITTS LKSEGEEEEE EEEEEEEEEG EGEEEEEEGE EEEEEGGEGE ELGEEEEVEE EGDVDSDEEE EEDEESSSEG LEAEDWAQGV VEAGGSFGAY GAQEEAQCPT LHFLEGGEDS DSDSEEEDDE EEDDEDEDDD DDEEDGDEVP VPSFGEAMAY FAMVKRYLTS FPIDDRVQSH ILHLEHDLVH VTRKNHARQA GVRGLGHQSP HHHHHH |
-Macromolecule #6: Human alpha-satellite DNA
| Macromolecule | Name: Human alpha-satellite DNA / type: dna / ID: 6 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GGAGGATTTC GTTGGAAACG GGATCAACTT CCCATAACTG AACGGAAGCA AACTCAGAAC ATTCTTTGTG ATGTTTGTAT TCAACTCACA GAGTTGAACC TTCCTTTGAT AGTTCAGGTT TGCAACACCC TTGTAGTAGA ATCTGCAAGT GTATATTTTG ACCACTTTGG A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)