-Search query
-Search result
Showing all 23 items for (author: stjepanovic & g)

EMDB-37752: 
Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation
Method: single particle / : Liu M, Wang Y, Su MY, Stjepanovic G

PDB-8wqr: 
Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation
Method: single particle / : Liu M, Wang Y, Su MY, Stjepanovic G

EMDB-37247: 
CULLIN3-KLHL22-RBX1 E3 ligase
Method: single particle / : Su MY
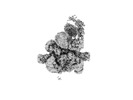
EMDB-37266: 
Cryo-EM structure of the KLHL22 E3 ligase bound to human glutamate dehydrogenase I
Method: single particle / : Su MY

PDB-8w4j: 
Cryo-EM structure of the KLHL22 E3 ligase bound to human glutamate dehydrogenase I
Method: single particle / : Su MY, Su MY

EMDB-37235: 
Human glutamate dehydrogenase I
Method: single particle / : Su MY

EMDB-34165: 
Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1
Method: single particle / : Cai Y, Zhang H

EMDB-34166: 
Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2
Method: single particle / : Cai Y, Zhang H
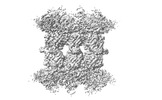
EMDB-34198: 
Human SARM1 bounded with NMN and Nanobody-C6, double-layer structure
Method: single particle / : Cai Y, Zhang H

PDB-8gni: 
Human SARM1 bounded with NMN and Nanobody-C6, Conformation 1
Method: single particle / : Cai Y, Zhang H

PDB-8gnj: 
Human SARM1 bounded with NMN and Nanobody-C6, Conformation 2
Method: single particle / : Cai Y, Zhang H

PDB-8gq5: 
Human SARM1 bounded with NMN and Nanobody-C6, double-layer structure
Method: single particle / : Cai Y, Zhang H

EMDB-8993: 
Class III PI3K Complex 2 with inhibitor Rubicon
Method: single particle / : Young LN, Morris KL, Hurley JH

EMDB-7337: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Basal state
Method: single particle / : Kasinath V, Faini M, Poepsel S, Reif D, Feng A, Stjepanovic G, Aebersold R, Nogales E

EMDB-7334: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State
Method: single particle / : Kasinath V, Faini M, Poepsel S, Reif D, Feng A, Stjepanovic G, Aebersold R, Nogales E

EMDB-7335: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State
Method: single particle / : Kasinath V, Faini M, Poepsel S, Reif D, Feng A, Stjepanovic G, Aebersold R, Nogales E

PDB-6c23: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Compact Active State
Method: single particle / : Kasinath V, Faini M, Poepsel S, Reif D, Feng A, Stjepanovic G, Aebersold R, Nogales E

PDB-6c24: 
Cryo-EM structure of PRC2 bound to cofactors AEBP2 and JARID2 in the Extended Active State
Method: single particle / : Kasinath V, Faini M, Poepsel S, Reif D, Feng A, Stjepanovic G, Aebersold R, Nogales E

EMDB-7005: 
Negative stain reconstruction of the peroxisomal AAA-ATPase Pex1/Pex6 complex associated with substrate Pex15
Method: single particle / : Chowdhury S, Gardner BM, Castanzo DT, Stjepanovic G, Stefely MS, Hurley JH, Martin A, Lander GC

EMDB-7072: 
RagA/RagC:Ragulator complex structure determined by single particle negative stain electron microscopy
Method: single particle / : Morris KL, Su M, Kim DJ, Fu Y, Lawrence R, Stjepanovic G, Zoncu R, Hurley JH

EMDB-2846: 
Three-dimensional structure of the autophagic phosphatidylinositol 3-kinase complex.
Method: single particle / : Baskaran S, Carlson LA, Stjepanovic G, Young LN, Kim DJ, Grob P, Stanley RE, Nogales E, Hurley JH
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search





 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
