+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6992 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the human PKD1/PKD2 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Su Q / Hu F / Ge X / Lei J / Yu S / Wang T / Zhou Q / Mei C / Shi Y | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Structure of the human PKD1-PKD2 complex. Authors: Qiang Su / Feizhuo Hu / Xiaofei Ge / Jianlin Lei / Shengqiang Yu / Tingliang Wang / Qiang Zhou / Changlin Mei / Yigong Shi /  Abstract: Mutations in two genes, and , account for most cases of autosomal dominant polycystic kidney disease, one of the most common monogenetic disorders. Here we report the 3.6-angstrom cryo-electron ...Mutations in two genes, and , account for most cases of autosomal dominant polycystic kidney disease, one of the most common monogenetic disorders. Here we report the 3.6-angstrom cryo-electron microscopy structure of truncated human PKD1-PKD2 complex assembled in a 1:3 ratio. PKD1 contains a voltage-gated ion channel (VGIC) fold that interacts with PKD2 to form the domain-swapped, yet noncanonical, transient receptor potential (TRP) channel architecture. The S6 helix in PKD1 is broken in the middle, with the extracellular half, S6a, resembling pore helix 1 in a typical TRP channel. Three positively charged, cavity-facing residues on S6b may block cation permeation. In addition to the VGIC, a five-transmembrane helix domain and a cytosolic PLAT domain were resolved in PKD1. The PKD1-PKD2 complex structure establishes a framework for dissecting the function and disease mechanisms of the PKD proteins. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6992.map.gz emd_6992.map.gz | 59.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6992-v30.xml emd-6992-v30.xml emd-6992.xml emd-6992.xml | 12.5 KB 12.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6992.png emd_6992.png | 238.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6992 http://ftp.pdbj.org/pub/emdb/structures/EMD-6992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6992 | HTTPS FTP |
-Validation report
| Summary document |  emd_6992_validation.pdf.gz emd_6992_validation.pdf.gz | 79.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6992_full_validation.pdf.gz emd_6992_full_validation.pdf.gz | 78.8 KB | Display | |
| Data in XML |  emd_6992_validation.xml.gz emd_6992_validation.xml.gz | 493 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6992 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6992 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_6992.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6992.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.091 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
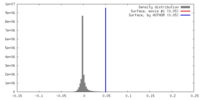
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : the structure of human PKD1/PKD2 complex
| Entire | Name: the structure of human PKD1/PKD2 complex |
|---|---|
| Components |
|
-Supramolecule #1: the structure of human PKD1/PKD2 complex
| Supramolecule | Name: the structure of human PKD1/PKD2 complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Samples were obtained by co-expression in 293F cells. A complex contains one PKD1 subunit and three PKD2 subunits. |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Mammalian expression vector pCK9 (others) / Recombinant cell: 293F cells |
-Macromolecule #1: human PKD2 (chain F,G)
| Macromolecule | Name: human PKD2 (chain F,G) / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Mammalian expression vector pCK9 (others) |
| Sequence | String: PRVAWAERLV RGLRGLWGTR LMEESSTNRE KYLKSVLREL VTYLLFLIVL CILTYGMMSS NVYYYTRMMS QLFLDTPVSK TEKTNFKTL SSMEDFWKFT EGSLLDGLYW KMQPSNQTEA DNRSFIFYEN LLLGVPRIRQ LRVRNGSCSI PQDLRDEIKE C YDVYSVSS ...String: PRVAWAERLV RGLRGLWGTR LMEESSTNRE KYLKSVLREL VTYLLFLIVL CILTYGMMSS NVYYYTRMMS QLFLDTPVSK TEKTNFKTL SSMEDFWKFT EGSLLDGLYW KMQPSNQTEA DNRSFIFYEN LLLGVPRIRQ LRVRNGSCSI PQDLRDEIKE C YDVYSVSS EDRAPFGPRN GTAWIYTSEK DLNGSSHWGI IATYSGAGYY LDLSRTREET AAQVASLKKN VWLDRGTRAT FI DFSVYNA NINLFCVVRL LVEFPATGGV IPSWQFQPLK LIRYVTTFDF FLAACEIIFC FFIFYYVVEE ILEIRIHKLH YFR SFWNCL DVVIVVLSVV AIGINIYRTS NVEVLLQFLE DQNTFPNFEH LAYWQIQFNN IAAVTVFFVW IKLFKFINFN RTMS QLSTT MSRCAKDLFG FAIMFFIIFL AYAQLAYLVF GTQVDDFSTF QECIFTQFRI ILGDINFAEI EEANRVLGPI YFTTF VFFM FFILLNMFLA IINDTYSEVK SDLAQQKAEM ELSDLIRKGY HKALVKLKLK KNTVD |
-Macromolecule #2: human PKD1 (chain B)
| Macromolecule | Name: human PKD1 (chain B) / type: protein_or_peptide / ID: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism: Mammalian expression vector pCK9 (others) |
| Sequence | String: TAFGASLFVP PSHVRFVFPE PTADVNYIVM LTCAVCLVTY MVMAAILHKL DQLDASRGRA IPFCGQRGRF KYEILVKTGW GRGSGTTAH VGIMLYGVDS RSGHRHLDGD RAFHRNSLDI FRIATPHSLG SVWKIRVWHD NKGLSPAWFL QHVIVRDLQT A RSAFFLVN ...String: TAFGASLFVP PSHVRFVFPE PTADVNYIVM LTCAVCLVTY MVMAAILHKL DQLDASRGRA IPFCGQRGRF KYEILVKTGW GRGSGTTAH VGIMLYGVDS RSGHRHLDGD RAFHRNSLDI FRIATPHSLG SVWKIRVWHD NKGLSPAWFL QHVIVRDLQT A RSAFFLVN DWLSVETEAN GGLVEKEVLA ASDAALLRFR RLLVAELQRG FFDKHIWLSI WDRPPRSRFT RIQRATCCVL LI CLFLGAN AVWYGAVGDS AYSTGHVSRL SPLSVDTVAV GLVSSVVVYP VYLAILFLFR MSRSKVAGSP SPTPAGQQVL DID SCLDSS VLDSSFLTFS GLHAEQAFVG QMKSDLFLDD SKSLVCWPSG EGTLSWPDLL SDPSIVGSNL RQLARGQAGH GLGP EEDGF SLASPYSPAK SFSASDEDLI QQVLAEGVSS PAPTQDTHME TDLLSSLSST PGEKTETLAL QRLGELGPPS PGLNW EQPQ AARLSRTGLV EGLRKRLLPA WCASLAHGLS LLLVAVAVAV SGWVGASFPP GVSVAWLLSS SASFLASFLG WEPLKV LLE ALYFSLVAKR LHPDEDDTLV ESPAVTPVSA RVPRVRPPHG FALFLAKEEA RKVKRLHGML RSLLVYMLFL LVTLLAS YG DASCHGHAYR LQSAIKQELH SRAFLAITRS EELWPWMAHV LLPYVHGNQS SPELGPPRLR QVRLQEALYP DPPGPRVH T CSAAGGFSTS DYDVGWESPH NGSGTWAYSA PDLLGAWSWG SCAVYDSGGY VQELGLSLEE SRDRLRFLQL HNWLDNRSR AVFLELTRYS PAVGLHAAVT LRLEFPAAGR ALAALSVRPF ALRRLSAGLS LPLLTSVCLL LFAVHFAVAE ARTWHREGRW RVLRLGAWA RWLLVALTAA TALVRLAQLG AADRQWTRFV RGRPRRFTSF DQVAQLSSAA RGLAASLLFL LLVKAAQQLR F VRQWSVFG KTLCRALPEL LGVTLGLVVL GVAYAQLAIL LVSSCVDSLW SVAQALLVLC PGTGLSTLCP AESWHLSPLL CV GLWALRL WGALRLGAVI LRWRYHALRG ELYRPAWEPQ DYEMVELFLR RLRLWMGLSK VKEFRHKVRF EGMEPLPSRS SRG S |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 281 K / Instrument: FEI VITROBOT MARK IV | ||||||||||||
| Details | This sample was monodisperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 116485 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)