+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-5691 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
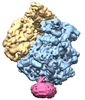
| タイトル | Structure of the SecY protein translocation channel in action | |||||||||
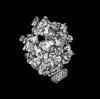
 マップデータ マップデータ | Reconstruction of archaeal 70S ribosome-SecYEbeta complex from Methanococcus jannaschii | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | archaeal 70S ribosome / SecYEbeta channel / co-translational translocation | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報intracellular protein transmembrane transport / SRP-dependent cotranslational protein targeting to membrane, translocation / signal sequence binding / protein secretion / protein transmembrane transporter activity / protein targeting / protein transport / plasma membrane 類似検索 - 分子機能 | |||||||||
| 生物種 |   Methanocaldococcus jannaschii (メタン生成菌) Methanocaldococcus jannaschii (メタン生成菌) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 9.0 Å | |||||||||
 データ登録者 データ登録者 | Park P / Menetret JF / Gumbart JC / Ludtke SJ / Li W / Whynot A / Rapoport TA / Akey CW | |||||||||
 引用 引用 |  ジャーナル: Nature / 年: 2014 ジャーナル: Nature / 年: 2014タイトル: Structure of the SecY channel during initiation of protein translocation. 著者: Eunyong Park / Jean-François Ménétret / James C Gumbart / Steven J Ludtke / Weikai Li / Andrew Whynot / Tom A Rapoport / Christopher W Akey /  要旨: Many secretory proteins are targeted by signal sequences to a protein-conducting channel, formed by prokaryotic SecY or eukaryotic Sec61 complexes, and are translocated across the membrane during ...Many secretory proteins are targeted by signal sequences to a protein-conducting channel, formed by prokaryotic SecY or eukaryotic Sec61 complexes, and are translocated across the membrane during their synthesis. Crystal structures of the inactive channel show that the SecY subunit of the heterotrimeric complex consists of two halves that form an hourglass-shaped pore with a constriction in the middle of the membrane and a lateral gate that faces the lipid phase. The closed channel has an empty cytoplasmic funnel and an extracellular funnel that is filled with a small helical domain, called the plug. During initiation of translocation, a ribosome-nascent chain complex binds to the SecY (or Sec61) complex, resulting in insertion of the nascent chain. However, the mechanism of channel opening during translocation is unclear. Here we have addressed this question by determining structures of inactive and active ribosome-channel complexes with cryo-electron microscopy. Non-translating ribosome-SecY channel complexes derived from Methanocaldococcus jannaschii or Escherichia coli show the channel in its closed state, and indicate that ribosome binding per se causes only minor changes. The structure of an active E. coli ribosome-channel complex demonstrates that the nascent chain opens the channel, causing mostly rigid body movements of the amino- and carboxy-terminal halves of SecY. In this early translocation intermediate, the polypeptide inserts as a loop into the SecY channel with the hydrophobic signal sequence intercalated into the open lateral gate. The nascent chain also forms a loop on the cytoplasmic surface of SecY rather than entering the channel directly. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_5691.map.gz emd_5691.map.gz | 1.2 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-5691-v30.xml emd-5691-v30.xml emd-5691.xml emd-5691.xml | 26.2 KB 26.2 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_5691.jpg emd_5691.jpg | 164.1 KB | ||
| マスクデータ |  emd_5691_msk_1.map emd_5691_msk_1.map emd_5691_msk_2.map emd_5691_msk_2.map emd_5691_msk_3.map emd_5691_msk_3.map emd_5691_msk_4.map emd_5691_msk_4.map emd_5691_msk_5.map emd_5691_msk_5.map | 16.4 MB 16.4 MB 160.8 KB 16.4 MB 2.2 MB |  マスクマップ マスクマップ | |
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5691 http://ftp.pdbj.org/pub/emdb/structures/EMD-5691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5691 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5691 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_5691_validation.pdf.gz emd_5691_validation.pdf.gz | 294.3 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_5691_full_validation.pdf.gz emd_5691_full_validation.pdf.gz | 293.9 KB | 表示 | |
| XML形式データ |  emd_5691_validation.xml.gz emd_5691_validation.xml.gz | 4.4 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5691 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5691 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5691 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_5691.map.gz / 形式: CCP4 / 大きさ: 16 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_5691.map.gz / 形式: CCP4 / 大きさ: 16 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Reconstruction of archaeal 70S ribosome-SecYEbeta complex from Methanococcus jannaschii | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 これらの図は立方格子座標系で作成されたものです | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 2.73 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
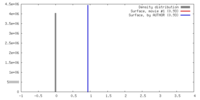
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
-セグメンテーションマップ: zone large ribosomal subunit
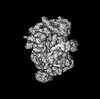
| 注釈 | zone large ribosomal subunit | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ファイル |  emd_5691_msk_1.map emd_5691_msk_1.map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
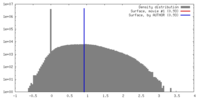
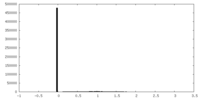
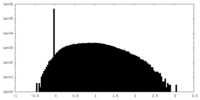
| 密度ヒストグラム |
-セグメンテーションマップ: zoned full channel desnity
| 注釈 | zoned full channel desnity | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ファイル |  emd_5691_msk_2.map emd_5691_msk_2.map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-セグメンテーションマップ: segmented density with only SecYEbeta
| 注釈 | segmented density with only SecYEbeta | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ファイル |  emd_5691_msk_3.map emd_5691_msk_3.map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-セグメンテーションマップ: zoned micelle
| 注釈 | zoned micelle | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ファイル |  emd_5691_msk_4.map emd_5691_msk_4.map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-セグメンテーションマップ: zoned small ribosomal subunit
| 注釈 | zoned small ribosomal subunit | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ファイル |  emd_5691_msk_5.map emd_5691_msk_5.map | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Methanococcus jannaschii 70S ribosome-SecYEbeta complex
| 全体 | 名称: Methanococcus jannaschii 70S ribosome-SecYEbeta complex |
|---|---|
| 要素 |
|
-超分子 #1000: Methanococcus jannaschii 70S ribosome-SecYEbeta complex
| 超分子 | 名称: Methanococcus jannaschii 70S ribosome-SecYEbeta complex タイプ: sample / ID: 1000 詳細: The sample was reconstituted from ribosomal subunits purified from M. jannaschii and from recombinant SecYEbeta. 集合状態: one 70S, one E-site tRNA, one SecYEbeta channel Number unique components: 3 |
|---|---|
| 分子量 | 理論値: 2.6 MDa |
-超分子 #1: non-translating 70S ribosome
| 超分子 | 名称: non-translating 70S ribosome / タイプ: complex / ID: 1 / 組換発現: No / データベース: NCBI / Ribosome-details: ribosome-prokaryote: ALL |
|---|---|
| 由来(天然) | 生物種:   Methanocaldococcus jannaschii (メタン生成菌) Methanocaldococcus jannaschii (メタン生成菌)細胞中の位置: cytoplasm |
| 分子量 | 理論値: 2.5 MDa |
-分子 #1: SecYEbeta
| 分子 | 名称: SecYEbeta / タイプ: protein_or_peptide / ID: 1 / Name.synonym: SecY channel / 詳細: SecY: Q60175.2, SecE: Q57817.1, Secbeta: P60460.1 / コピー数: 1 / 集合状態: heterotrimer / 組換発現: Yes |
|---|---|
| 由来(天然) | 生物種:   Methanocaldococcus jannaschii (メタン生成菌) Methanocaldococcus jannaschii (メタン生成菌)細胞中の位置: inner membrane |
| 分子量 | 理論値: 61.8 KDa |
| 組換発現 | 生物種:  |
-分子 #2: transfer RNA
| 分子 | 名称: transfer RNA / タイプ: rna / ID: 2 / Name.synonym: tRNA / 詳細: co-purified with the ribosomal subunits / 分類: TRANSFER / Structure: DOUBLE HELIX / Synthetic?: No |
|---|---|
| 由来(天然) | 生物種:   Methanocaldococcus jannaschii (メタン生成菌) Methanocaldococcus jannaschii (メタン生成菌) |
| 分子量 | 理論値: 25 KDa |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 2 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 詳細: 100 mM NH4Cl, 30 mM MgCl2, 20 mM HEPES-KOH, 6 mM beta-mercaptoethanol, 0.1% DDM |
| グリッド | 詳細: Quantifoil 400 mesh 2/1 Cu grids, air glow discharged, and 400 mesh Cu grids with a thin continuous carbon foil |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 77 K / 装置: FEI VITROBOT MARK III / 手法: Blot 1-2 seconds before plunging. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TECNAI F20 |
|---|---|
| 温度 | 平均: 93 K |
| アライメント法 | Legacy - 非点収差: corrected at 175,000 times magnification |
| 詳細 | Low dose imaging, data collected manually |
| 日付 | 2008年4月10日 |
| 撮影 | カテゴリ: FILM / フィルム・検出器のモデル: KODAK SO-163 FILM / デジタル化 - スキャナー: OTHER / 実像数: 217 / 平均電子線量: 20 e/Å2 / 詳細: Zeiss SCAI scanner was also used. / Od range: 1 / ビット/ピクセル: 16 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 倍率(補正後): 51000 / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / Cs: 2.0 mm / 最大 デフォーカス(公称値): 2.5 µm / 最小 デフォーカス(公称値): 1.0 µm / 倍率(公称値): 50000 |
| 試料ステージ | 試料ホルダー: Oxford holder / 試料ホルダーモデル: GATAN LIQUID NITROGEN |
| 実験機器 |  モデル: Tecnai F20 / 画像提供: FEI Company |
- 画像解析
画像解析
| 詳細 | Particles were selected with e2boxer and CTF-corrected with EMAN2. Final maps were obtained from 6 different refinement conditions, aligned in Chimera, and averaged to produce the final map. |
|---|---|
| CTF補正 | 詳細: per micrograph |
| 最終 再構成 | アルゴリズム: OTHER / 解像度のタイプ: BY AUTHOR / 解像度: 9.0 Å / 解像度の算出法: OTHER / ソフトウェア - 名称: EMAN1, EMAN2 詳細: Final map calculated as an average of 6 different refinements in EMAN2 with different parameters. 使用した粒子像数: 37000 |
| 最終 2次元分類 | クラス数: 3800 |
-原子モデル構築 1
| 初期モデル | PDB ID:  3j21 |
|---|---|
| ソフトウェア | 名称: Chimera, MDFF |
| 詳細 | Docked with Chimera and fit with MDFF. Chains omitted: 1('el' 1-77), 6(L83-1), 4(L83-2), c(L33e). Chains truncated: a(L31e) 78-85, W(L29) 67-72, C(L3) 103-125. |
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT |
| 得られたモデル |  PDB-4v4n: |
-原子モデル構築 2
| 初期モデル | PDB ID:  3j2l |
|---|---|
| ソフトウェア | 名称: Chimera, MDFF |
| 詳細 | Regions omitted: rRNA chain 1 [1-6, 3047-3049 5' and 3' ends 23S], 150-163, 750-756, 1311-1321, 2904-2942 (ES). |
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT |
| 得られたモデル |  PDB-4v4n: |
-原子モデル構築 3
| 初期モデル | PDB ID:  3j20 |
|---|---|
| ソフトウェア | 名称: Chimera, MDFF |
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT |
| 得られたモデル |  PDB-4v4n: |
 ムービー
ムービー コントローラー
コントローラー

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)