+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4575 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Full length GluA1/2-gamma8 complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationPhase 0 - rapid depolarisation / Phase 2 - plateau phase / Cargo concentration in the ER / cellular response to amine stimulus / axonal spine / COPII-mediated vesicle transport / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / cellular response to ammonium ion / response to sucrose ...Phase 0 - rapid depolarisation / Phase 2 - plateau phase / Cargo concentration in the ER / cellular response to amine stimulus / axonal spine / COPII-mediated vesicle transport / positive regulation of locomotion involved in locomotory behavior / positive regulation of membrane potential / cellular response to ammonium ion / response to sucrose / proximal dendrite / neurotransmitter receptor transport, postsynaptic endosome to lysosome / L-type voltage-gated calcium channel complex / LGI-ADAM interactions / myosin V binding / Trafficking of AMPA receptors / neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration / neuron spine / cellular response to L-glutamate / regulation of AMPA receptor activity / conditioned place preference / neurotransmitter receptor internalization / protein phosphatase 2B binding / response to arsenic-containing substance / regulation of monoatomic ion transmembrane transport / postsynaptic neurotransmitter receptor diffusion trapping / cellular response to dsRNA / dendritic spine membrane / Synaptic adhesion-like molecules / long-term synaptic depression / beta-2 adrenergic receptor binding / cellular response to peptide hormone stimulus / response to morphine / protein kinase A binding / peptide hormone receptor binding / spine synapse / response to psychosocial stress / dendritic spine neck / spinal cord development / dendritic spine head / Activation of AMPA receptors / neuronal cell body membrane / perisynaptic space / AMPA glutamate receptor activity / transmission of nerve impulse / ligand-gated monoatomic cation channel activity / channel regulator activity / Trafficking of GluR2-containing AMPA receptors / response to lithium ion / immunoglobulin binding / behavioral response to pain / regulation of postsynaptic membrane neurotransmitter receptor levels / AMPA glutamate receptor complex / kainate selective glutamate receptor activity / adenylate cyclase binding / ionotropic glutamate receptor complex / extracellularly glutamate-gated ion channel activity / positive regulation of excitatory postsynaptic potential / cellular response to organic cyclic compound / cellular response to glycine / asymmetric synapse / excitatory synapse / calcium channel regulator activity / regulation of receptor recycling / neuronal action potential / G-protein alpha-subunit binding / regulation of postsynaptic membrane potential / Unblocking of NMDA receptors, glutamate binding and activation / postsynaptic density, intracellular component / voltage-gated calcium channel activity / positive regulation of synaptic transmission / glutamate receptor binding / extracellular ligand-gated monoatomic ion channel activity / long-term memory / response to electrical stimulus / glutamate-gated receptor activity / response to fungicide / glutamate-gated calcium ion channel activity / presynaptic active zone membrane / regulation of synaptic transmission, glutamatergic / somatodendritic compartment / dendrite membrane / synapse assembly / cellular response to brain-derived neurotrophic factor stimulus / cytoskeletal protein binding / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / ionotropic glutamate receptor binding / dendrite cytoplasm / ionotropic glutamate receptor signaling pathway / positive regulation of synaptic transmission, glutamatergic / response to nutrient levels / SNARE binding / dendritic shaft / synaptic transmission, glutamatergic / synaptic membrane / response to cocaine / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / PDZ domain binding / long-term synaptic potentiation / protein tetramerization Similarity search - Function | |||||||||
| Biological species |  | |||||||||
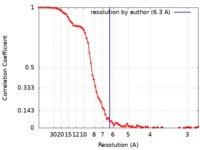
| Method | single particle reconstruction / cryo EM / Resolution: 6.3 Å | |||||||||
 Authors Authors | Herguedas B / Garcia-Nafria J / Greger IG | |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Architecture of the heteromeric GluA1/2 AMPA receptor in complex with the auxiliary subunit TARP γ8. Authors: Beatriz Herguedas / Jake F Watson / Hinze Ho / Ondrej Cais / Javier García-Nafría / Ingo H Greger /  Abstract: AMPA-type glutamate receptors (AMPARs) mediate excitatory neurotransmission and are central regulators of synaptic plasticity, a molecular mechanism underlying learning and memory. Although AMPARs ...AMPA-type glutamate receptors (AMPARs) mediate excitatory neurotransmission and are central regulators of synaptic plasticity, a molecular mechanism underlying learning and memory. Although AMPARs act predominantly as heteromers, structural studies have focused on homomeric assemblies. Here, we present a cryo-electron microscopy structure of the heteromeric GluA1/2 receptor associated with two transmembrane AMPAR regulatory protein (TARP) γ8 auxiliary subunits, the principal AMPAR complex at hippocampal synapses. Within the receptor, the core subunits arrange to give the GluA2 subunit dominant control of gating. This structure reveals the geometry of the Q/R site that controls calcium flux, suggests association of TARP-stabilized lipids, and demonstrates that the extracellular loop of γ8 modulates gating by selectively interacting with the GluA2 ligand-binding domain. Collectively, this structure provides a blueprint for deciphering the signal transduction mechanisms of synaptic AMPARs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4575.map.gz emd_4575.map.gz | 6.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4575-v30.xml emd-4575-v30.xml emd-4575.xml emd-4575.xml | 18.1 KB 18.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4575_fsc.xml emd_4575_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4575.png emd_4575.png | 65.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4575 http://ftp.pdbj.org/pub/emdb/structures/EMD-4575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4575 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4575 | HTTPS FTP |
-Validation report
| Summary document |  emd_4575_validation.pdf.gz emd_4575_validation.pdf.gz | 247.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4575_full_validation.pdf.gz emd_4575_full_validation.pdf.gz | 246.6 KB | Display | |
| Data in XML |  emd_4575_validation.xml.gz emd_4575_validation.xml.gz | 9.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4575 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4575 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4575 | HTTPS FTP |
-Related structure data
| Related structure data |  6qkzMC  4572C  6qkcC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_4575.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4575.map.gz / Format: CCP4 / Size: 40.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.4 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
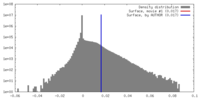
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : GluA1/A2 bound to gamma-8
| Entire | Name: GluA1/A2 bound to gamma-8 |
|---|---|
| Components |
|
-Supramolecule #1: GluA1/A2 bound to gamma-8
| Supramolecule | Name: GluA1/A2 bound to gamma-8 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 447 kDa/nm |
-Macromolecule #1: GluA1
| Macromolecule | Name: GluA1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 100.739602 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ADYKDDDDKN FPNNIQIGGL FPNQQSQEHA AFRFALSQLT EPPKLLPQID IVNISDSFEM TYRFCSQFSK GVYAIFGFYE RRTVNMLTS FCGALHVCFI TPSFPVDTSN QFVLQLRPEL QEALISIIDH YKWQTFVYIY DADRGLSVLQ RVLDTAAEKN W QVTAVNIL ...String: ADYKDDDDKN FPNNIQIGGL FPNQQSQEHA AFRFALSQLT EPPKLLPQID IVNISDSFEM TYRFCSQFSK GVYAIFGFYE RRTVNMLTS FCGALHVCFI TPSFPVDTSN QFVLQLRPEL QEALISIIDH YKWQTFVYIY DADRGLSVLQ RVLDTAAEKN W QVTAVNIL TTTEEGYRML FQDLEKKKER LVVVDCESER LNAILGQIVK LEKNGIGYHY ILANLGFMDI DLNKFKESGA NV TGFQLVN YTDTIPARIM QQWRTSDSRD HTRVDWKRPK YTSALTYDGV KVMAEAFQSL RRQRIDISRR GNAGDCLANP AVP WGQGID IQRALQQVRF EGLTGNVQFN EKGRRTNYTL HVIEMKHDGI RKIGYWNEDD KFVPAATDAQ AGGDNSSVQN RTYI VTTIL EDPYVMLKKN ANQFEGNDRY EGYCVELAAE IAKHVGYSYR LEIVSDGKYG ARDPDTKAWN GMVGELVYGR ADVAV APLT ITLVREEVID FSKPFMSLGI SIMIKKPQKS KPGVFSFLDP LAYEIWMCIV FAYIGVSVVL FLVSRFSPYE WHSEEF EEG RDQTTSDQSN EFGIFNSLWF SLGAFMQQGC DISPRSLSGR IVGGVWWFFT LIIISSYTAN LAAFLTVERM VSPIESA ED LAKQTEIAYG TLEAGSTKEF FRRSKIAVFE KMWTYMKSAE PSVFVRTTEE GMIRVRKSKG KYAYLLESTM NEYIEQRK P CDTMKVGGNL DSKGYGIATP KGSALRGPVN LAVLKLSEQG VLDKLKSKWW YDKGECGSKD SGSKDKTSAL SLSNVAGVF YILIGGLGLA MLVALIEFCY KSRSESKRMK GFCLIPQQSI NEAIRTSTLP RNSGAGASGG GGSGENGRVV SQDFPKSMQS IPCMSHSSG MPLGATGL |
-Macromolecule #2: Glutamate receptor 2
| Macromolecule | Name: Glutamate receptor 2 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 93.880078 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VSSNSIQIGG LFPRGADQEY SAFRVGMVQF STSEFRLTPH IDNLEVANSF AVTNAFCSQF SRGVYAIFGF YDKKSVNTIT SFCGTLHVS FITPSFPTDG THPFVIQMRP DLKGALLSLI EYYQWDKFAY LYDSDRGLST LQAVLDSAAE KKWQVTAINV G NINNDKKD ...String: VSSNSIQIGG LFPRGADQEY SAFRVGMVQF STSEFRLTPH IDNLEVANSF AVTNAFCSQF SRGVYAIFGF YDKKSVNTIT SFCGTLHVS FITPSFPTDG THPFVIQMRP DLKGALLSLI EYYQWDKFAY LYDSDRGLST LQAVLDSAAE KKWQVTAINV G NINNDKKD ETYRSLFQDL ELKKERRVIL DCERDKVNDI VDQVITIGKH VKGYHYIIAN LGFTDGDLLK IQFGGANVSG FQ IVDYDDS LVSKFIERWS TLEEKEYPGA HTATIKYTSA LTYDAVQVMT EAFRNLRKQR IEISRRGNAG DCLANPAVPW GQG VEIERA LKQVQVEGLS GNIKFDQNGK RINYTINIME LKTNGPRKIG YWSEVDKMVV TLTELPSGND TSGLENKTVV VTTI LESPY VMMKKNHEML EGNERYEGYC VDLAAEIAKH CGFKYKLTIV GDGKYGARDA DTKIWNGMVG ELVYGKADIA IAPLT ITLV REEVIDFSKP FMSLGISIMI KKPQKSKPGV FSFLDPLAYE IWMCIVFAYI GVSVVLFLVS RFSPYEWHTE EFEDGR ETQ SSESTNEFGI FNSLWFSLGA FMRQGCDISP RSLSGRIVGG VWWFFTLIII SSYTANLAAF LTVERMVSPI ESAEDLS KQ TEIAYGTLDS GSTKEFFRRS KIAVFDKMWT YMRSAEPSVF VRTTAEGVAR VRKSKGKYAY LLESTMNEYI EQRKPCDT M KVGGNLDSKG YGIATPKGSS LGTPVNLAVL KLSEQGVLDK LKNKWWYDKG ECGAKDSGSK EKTSALSLSN VAGVFYILV GGLGLAMLVA LIEFCYKSRA EAKRMKVAKN PQNINPSSS |
-Macromolecule #3: Voltage-dependent calcium channel gamma-8 subunit
| Macromolecule | Name: Voltage-dependent calcium channel gamma-8 subunit / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 43.592008 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GESLKRWNEE RGLWCEKGVQ VLLTTIGAFA AFGLMTIAIS TDYWLYTRAL ICNTTNLTAG DDGPPHRGGS GSSEKKDPGG LTHSGLWRI CCLEGLKRGV CVKINHFPED TDYDHDSSEY LLRVVRASSI FPILSAILLL LGGVCVAASR VYKSKRNIIL G AGILFVAA ...String: GESLKRWNEE RGLWCEKGVQ VLLTTIGAFA AFGLMTIAIS TDYWLYTRAL ICNTTNLTAG DDGPPHRGGS GSSEKKDPGG LTHSGLWRI CCLEGLKRGV CVKINHFPED TDYDHDSSEY LLRVVRASSI FPILSAILLL LGGVCVAASR VYKSKRNIIL G AGILFVAA GLSNIIGVIV YISANAGEPG PKRDEEKKNH YSYGWSFYFG GLSFILAEVI GVLAVNIYIE RSREAHCQSR SD LLKAGGG AGGSGGSGPS AILRLPSYRF RYRRRSRSSS RGSSEASPSR DASPGGPGGP GFASTDISMY TLSRDPSKGS VAA GLASAG GGGGGAGVGA YGGAAGAAGG GGTGSERDRG SSAGFLTLHN AFPKEAASGV TVTVTGPPAA PAPAPPAPAA PAPG TLSKE AAASNTNTLN RKLEVLFQ |
-Macromolecule #6: 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-s...
| Macromolecule | Name: 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide type: ligand / ID: 6 / Number of copies: 4 / Formula: E2Q |
|---|---|
| Molecular weight | Theoretical: 336.28 Da |
| Chemical component information | 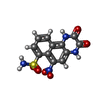 ChemComp-E2Q: |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 25 mM TRIS, pH 8, 150 mM NaCl and 0.1 % digitonin (w/v) |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: 3uL on grid, 60 sec incubation and 4sec blotting time. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Specialist optics | Phase plate: VOLTA PHASE PLATE / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Number grids imaged: 3 / Number real images: 5005 / Average exposure time: 14.0 sec. / Average electron dose: 32.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -0.9 µm / Nominal defocus min: -0.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)