+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3944 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | BK polyomavirus + 20 mM GT1b oligosaccharide | |||||||||
 Map data Map data | BK polyomavirus in complex with GT1b oligosaccharide. | |||||||||
 Sample Sample | BK polyomavirus != Human polyomavirus 1 BK polyomavirus
| |||||||||
 Keywords Keywords | Polyomavirus / Receptor / Complex / Glycan / VIRUS | |||||||||
| Function / homology |  Function and homology information Function and homology informationcaveolin-mediated endocytosis of virus by host cell / symbiont entry into host cell via permeabilization of host membrane / T=7 icosahedral viral capsid / viral penetration into host nucleus / viral capsid / host cell / channel activity / monoatomic ion transmembrane transport / host cell endoplasmic reticulum membrane / endocytosis involved in viral entry into host cell ...caveolin-mediated endocytosis of virus by host cell / symbiont entry into host cell via permeabilization of host membrane / T=7 icosahedral viral capsid / viral penetration into host nucleus / viral capsid / host cell / channel activity / monoatomic ion transmembrane transport / host cell endoplasmic reticulum membrane / endocytosis involved in viral entry into host cell / virion attachment to host cell / host cell nucleus / structural molecule activity / DNA binding / membrane Similarity search - Function | |||||||||
| Biological species |  BK polyomavirus / BK polyomavirus /  Human polyomavirus 1 Human polyomavirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Hurdiss DL / Ranson NA | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2018 Journal: Structure / Year: 2018Title: The Structure of an Infectious Human Polyomavirus and Its Interactions with Cellular Receptors. Authors: Daniel L Hurdiss / Martin Frank / Joseph S Snowden / Andrew Macdonald / Neil A Ranson /   Abstract: BK polyomavirus (BKV) causes polyomavirus-associated nephropathy and hemorrhagic cystitis in immunosuppressed patients. These are diseases for which we currently have limited treatment options, but ...BK polyomavirus (BKV) causes polyomavirus-associated nephropathy and hemorrhagic cystitis in immunosuppressed patients. These are diseases for which we currently have limited treatment options, but potential therapies could include pre-transplant vaccination with a multivalent BKV vaccine or therapeutics which inhibit capsid assembly or block attachment and entry into target cells. A useful tool in such efforts would be a high-resolution structure of the infectious BKV virion and how this interacts with its full repertoire of cellular receptors. We present the 3.4-Å cryoelectron microscopy structure of native, infectious BKV in complex with the receptor fragment of GT1b ganglioside. We also present structural evidence that BKV can utilize glycosaminoglycans as attachment receptors. This work highlights features that underpin capsid stability and provides a platform for rational design and development of urgently needed pharmacological interventions for BKV-associated diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3944.map.gz emd_3944.map.gz | 452.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3944-v30.xml emd-3944-v30.xml emd-3944.xml emd-3944.xml | 13.4 KB 13.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3944_fsc.xml emd_3944_fsc.xml | 20.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_3944.png emd_3944.png | 221.8 KB | ||
| Filedesc metadata |  emd-3944.cif.gz emd-3944.cif.gz | 5.5 KB | ||
| Others |  emd_3944_additional.map.gz emd_3944_additional.map.gz | 656.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3944 http://ftp.pdbj.org/pub/emdb/structures/EMD-3944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3944 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3944 | HTTPS FTP |
-Validation report
| Summary document |  emd_3944_validation.pdf.gz emd_3944_validation.pdf.gz | 327.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3944_full_validation.pdf.gz emd_3944_full_validation.pdf.gz | 326.3 KB | Display | |
| Data in XML |  emd_3944_validation.xml.gz emd_3944_validation.xml.gz | 17.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3944 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3944 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3944 | HTTPS FTP |
-Related structure data
| Related structure data |  6esbMC  3945C  3946C  4212C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3944.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3944.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BK polyomavirus in complex with GT1b oligosaccharide. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0651 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
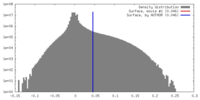
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: BK polyomavirus in complex with GT1b oligosaccharide (unsharpened...
| File | emd_3944_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | BK polyomavirus in complex with GT1b oligosaccharide (unsharpened map). | ||||||||||||
| Projections & Slices |
| ||||||||||||
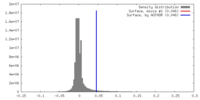
| Density Histograms |
- Sample components
Sample components
-Entire : BK polyomavirus
| Entire | Name:  BK polyomavirus BK polyomavirus |
|---|---|
| Components |
|
-Supramolecule #1: Human polyomavirus 1
| Supramolecule | Name: Human polyomavirus 1 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / NCBI-ID: 1891762 / Sci species name: Human polyomavirus 1 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Virus shell | Shell ID: 1 / Name: Icosahedron / Diameter: 500.0 Å / T number (triangulation number): 7 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  BK polyomavirus BK polyomavirus |
| Molecular weight | Theoretical: 40.053449 KDa |
| Sequence | String: APTKRKGECP GAAPKKPKEP VQVPKLLIKG GVEVLEVKTG VDAITEVECF LNPEMGDPDE NLRGFSLKLS AENDFSSDSP ERKMLPCYS TARIPLPNLN EDLTCGNLLM WEAVTVQTEV IGITSMLNLH AGSQKVHEHG GGKPIQGSNF HFFAVGGDPL E MQGVLMNY ...String: APTKRKGECP GAAPKKPKEP VQVPKLLIKG GVEVLEVKTG VDAITEVECF LNPEMGDPDE NLRGFSLKLS AENDFSSDSP ERKMLPCYS TARIPLPNLN EDLTCGNLLM WEAVTVQTEV IGITSMLNLH AGSQKVHEHG GGKPIQGSNF HFFAVGGDPL E MQGVLMNY RTKYPDGTIT PKNPTAQSQV MNTDHKAYLD KNNAYPVECW VPDPSRNENT RYFGTFTGGE NVPPVLHVTN TA TTVLLDE QGVGPLCKAD SLYVSAADIC GLFTNSSGTQ QWRGLARYFK IRLRKRSVKN PYPISFLLSD LINRRTQRVD GQP MYGMES QVEEVRVFDG TERLPGDPDM IRYIDKQGQL QTKML UniProtKB: Capsid protein VP1 |
-Macromolecule #2: Minor capsid protein VP2
| Macromolecule | Name: Minor capsid protein VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  BK polyomavirus BK polyomavirus |
| Molecular weight | Theoretical: 38.25273 KDa |
| Sequence | String: GAALALLGDL VASVSEAAAA TGFSVAEIAA GEAAAAIEVQ IASLATVEGI TSTSEAIAAI GLTPQTYAVI AGAPGAIAGF AALIQTVSG ISSLAQVGYR FFSDWDHKVS TVGLYQQSGM ALELFNPDEY YDILFPGVNT FVNNIQYLDP RHWGPSLFAT I SQALWHVI ...String: GAALALLGDL VASVSEAAAA TGFSVAEIAA GEAAAAIEVQ IASLATVEGI TSTSEAIAAI GLTPQTYAVI AGAPGAIAGF AALIQTVSG ISSLAQVGYR FFSDWDHKVS TVGLYQQSGM ALELFNPDEY YDILFPGVNT FVNNIQYLDP RHWGPSLFAT I SQALWHVI RDDIPSITSQ ELQRRTERFF RDSLARFLEE TTWTIVNAPI NFYNYIQQYY SDLSPIRPSM VRQVAEREGT RV HFGHTYS IDDADSIEEV TQRMDLRNQQ SVHSGEFIEK TIAPGGANQR TAPQWMLPLL LGLYGTVTPA LEAYEDGPNQ KKR RVSRGS SQKAKGTRAS AKTTNKRRSR SSRS UniProtKB: Minor capsid protein VP2 |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 6 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.25 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)