[English] 日本語
 Yorodumi
Yorodumi- EMDB-3425: Electron cryo-microscopy of human P-glycoprotein: ATP bound - clo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3425 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Electron cryo-microscopy of human P-glycoprotein: ATP bound - closed conformation | |||||||||
 Map data Map data | Reconstruction of mutant (E556Q/E1201Q) human P-glycoprotein in an ATP binding state, bound to UIC2 Fab. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | human P-Glycoprotein / ABCB1 / MDR-1 / closed conformation / ATP bound / UIC2 Fab | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of anion channel activity / carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / terpenoid transport / ceramide floppase activity / regulation of response to osmotic stress / floppase activity / ceramide translocation / Abacavir transmembrane transport / external side of apical plasma membrane ...positive regulation of anion channel activity / carboxylic acid transmembrane transport / carboxylic acid transmembrane transporter activity / terpenoid transport / ceramide floppase activity / regulation of response to osmotic stress / floppase activity / ceramide translocation / Abacavir transmembrane transport / external side of apical plasma membrane / Atorvastatin ADME / phosphatidylethanolamine flippase activity / xenobiotic transport across blood-brain barrier / transepithelial transport / phosphatidylcholine floppase activity / xenobiotic detoxification by transmembrane export across the plasma membrane / export across plasma membrane / ABC-type xenobiotic transporter / P-type phospholipid transporter / ABC-type xenobiotic transporter activity / phospholipid translocation / Prednisone ADME / efflux transmembrane transporter activity / xenobiotic transmembrane transporter activity / transmembrane transporter activity / ATPase-coupled transmembrane transporter activity / transport across blood-brain barrier / xenobiotic metabolic process / regulation of chloride transport / stem cell proliferation / ABC-family proteins mediated transport / transmembrane transport / G2/M transition of mitotic cell cycle / response to xenobiotic stimulus / apical plasma membrane / ubiquitin protein ligase binding / cell surface / ATP hydrolysis activity / extracellular exosome / ATP binding / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 19.0 Å | |||||||||
 Authors Authors | Frank GA / Shukla S / Rao P / Borgnia MJ / Bartesaghi A / Merk A / Mobin A / Esser L / Earl LA / Gottesman MM ...Frank GA / Shukla S / Rao P / Borgnia MJ / Bartesaghi A / Merk A / Mobin A / Esser L / Earl LA / Gottesman MM / Xia D / Ambudkar SV / Subramaniam S | |||||||||
 Citation Citation |  Journal: Mol Pharmacol / Year: 2016 Journal: Mol Pharmacol / Year: 2016Title: Cryo-EM Analysis of the Conformational Landscape of Human P-glycoprotein (ABCB1) During its Catalytic Cycle. Authors: Gabriel A Frank / Suneet Shukla / Prashant Rao / Mario J Borgnia / Alberto Bartesaghi / Alan Merk / Aerfa Mobin / Lothar Esser / Lesley A Earl / Michael M Gottesman / Di Xia / Suresh V ...Authors: Gabriel A Frank / Suneet Shukla / Prashant Rao / Mario J Borgnia / Alberto Bartesaghi / Alan Merk / Aerfa Mobin / Lothar Esser / Lesley A Earl / Michael M Gottesman / Di Xia / Suresh V Ambudkar / Sriram Subramaniam /  Abstract: The multidrug transporter P-glycoprotein (P-gp, ABCB1) is an ATP-dependent pump that mediates the efflux of structurally diverse drugs and xenobiotics across cell membranes, affecting drug ...The multidrug transporter P-glycoprotein (P-gp, ABCB1) is an ATP-dependent pump that mediates the efflux of structurally diverse drugs and xenobiotics across cell membranes, affecting drug pharmacokinetics and contributing to the development of multidrug resistance. Structural information about the conformational changes in human P-gp during the ATP hydrolysis cycle has not been directly demonstrated, although mechanistic information has been inferred from biochemical and biophysical studies conducted with P-gp and its orthologs, or from structures of other ATP-binding cassette transporters. Using single-particle cryo-electron microscopy, we report the surprising discovery that, in the absence of the transport substrate and nucleotides, human P-gp can exist in both open [nucleotide binding domains (NBDs) apart; inward-facing] and closed (NBDs close; outward-facing) conformations. We also probe conformational states of human P-gp during the catalytic cycle, and demonstrate that, following ATP hydrolysis, P-gp transitions through a complete closed conformation to a complete open conformation in the presence of ADP. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3425.map.gz emd_3425.map.gz | 9.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3425-v30.xml emd-3425-v30.xml emd-3425.xml emd-3425.xml | 13 KB 13 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3425.png emd_3425.png | 1002 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3425 http://ftp.pdbj.org/pub/emdb/structures/EMD-3425 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3425 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3425 | HTTPS FTP |
-Validation report
| Summary document |  emd_3425_validation.pdf.gz emd_3425_validation.pdf.gz | 189.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3425_full_validation.pdf.gz emd_3425_full_validation.pdf.gz | 189 KB | Display | |
| Data in XML |  emd_3425_validation.xml.gz emd_3425_validation.xml.gz | 6.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3425 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3425 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3425 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3425 | HTTPS FTP |
-Related structure data
| Related structure data |  3421C  3422C  3423C  3424C  3426C  3427C  3428C  3429C  3430C C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3425.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3425.map.gz / Format: CCP4 / Size: 41.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of mutant (E556Q/E1201Q) human P-glycoprotein in an ATP binding state, bound to UIC2 Fab. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.403 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
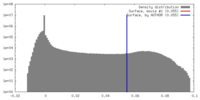
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human P-glycoprotein (E556Q/E1201Q) in ATP-binding, bound to UIC2 Fab
| Entire | Name: Human P-glycoprotein (E556Q/E1201Q) in ATP-binding, bound to UIC2 Fab |
|---|---|
| Components |
|
-Supramolecule #1000: Human P-glycoprotein (E556Q/E1201Q) in ATP-binding, bound to UIC2 Fab
| Supramolecule | Name: Human P-glycoprotein (E556Q/E1201Q) in ATP-binding, bound to UIC2 Fab type: sample / ID: 1000 Oligomeric state: One monomer of P-glycoprotein bound to one UIC2 Fab Number unique components: 2 |
|---|---|
| Molecular weight | Theoretical: 190 KDa |
-Macromolecule #1: P-glycoprotein
| Macromolecule | Name: P-glycoprotein / type: protein_or_peptide / ID: 1 / Name.synonym: P-gp, Pgp, ABCB1, MDR1 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) / synonym: Human / Location in cell: Plasma membrane Homo sapiens (human) / synonym: Human / Location in cell: Plasma membrane |
| Molecular weight | Theoretical: 141 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) / Recombinant strain: BTI-TN-5B1-4 / Recombinant cell: High Five insect cells Trichoplusia ni (cabbage looper) / Recombinant strain: BTI-TN-5B1-4 / Recombinant cell: High Five insect cells |
| Sequence | UniProtKB: ATP-dependent translocase ABCB1 InterPro: AAA+ ATPase domain, ABC transporter type 1, transmembrane domain, ABC transporter-like, ATP-binding domain, ABC transporter-like, conserved site, P-loop containing nucleoside triphosphate hydrolase |
-Macromolecule #2: UIC2 Fab
| Macromolecule | Name: UIC2 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Oligomeric state: Monomer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 50 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: Hybridoma Homo sapiens (human) / Recombinant cell: Hybridoma |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7.5 Details: 10 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.09% n-Dodecyl Betta-D-maltoside |
| Grid | Details: 200 mesh Cu R1.2/1.3 holey carbon grids from Quantifoil, plasma-cleaned |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 83 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Cs | 0 |
| Date | Nov 19, 2014 |
| Image recording | Category: CCD / Film or detector model: FEI FALCON II (4k x 4k) / Number real images: 2027 / Average electron dose: 30 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 47000 / Cs: mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | he particles were selected using an automatic selection program. |
|---|---|
| CTF correction | Details: CTF parameters obtained from whole micrograph |
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 19.0 Å / Resolution method: OTHER / Software - Name: EMAN2, Relion, 1.3 / Number images used: 14158 |
| Final two d classification | Number classes: 5 |
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Overlap |
-Atomic model buiding 2
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Overlap |
-Atomic model buiding 3
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Overlap |
-Atomic model buiding 4
| Initial model | PDB ID: |
|---|---|
| Software | Name:  Chimera Chimera |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Overlap |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)