[English] 日本語
 Yorodumi
Yorodumi- EMDB-30686: Krios G4 apoferritin test with K3/(slit out) SerialEM BIS 1x1x4 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30686 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Krios G4 apoferritin test with K3/(slit out) SerialEM BIS 1x1x4 | |||||||||
 Map data Map data | Masked apoferritin map | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationIron uptake and transport / Golgi Associated Vesicle Biogenesis / iron ion sequestering activity / negative regulation of ferroptosis / ferroxidase / autolysosome / ferroxidase activity / intracellular sequestering of iron ion / negative regulation of fibroblast proliferation / endocytic vesicle lumen ...Iron uptake and transport / Golgi Associated Vesicle Biogenesis / iron ion sequestering activity / negative regulation of ferroptosis / ferroxidase / autolysosome / ferroxidase activity / intracellular sequestering of iron ion / negative regulation of fibroblast proliferation / endocytic vesicle lumen / autophagosome / Neutrophil degranulation / ferric iron binding / ferrous iron binding / iron ion transport / immune response / iron ion binding / negative regulation of cell population proliferation / mitochondrion / extracellular region / identical protein binding / membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
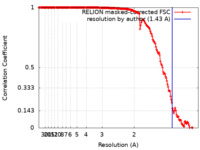
| Method | single particle reconstruction / cryo EM / Resolution: 1.43 Å | |||||||||
 Authors Authors | Danev R | |||||||||
| Funding support |  Japan, 1 items Japan, 1 items
| |||||||||
 Citation Citation |  Journal: Microscopy (Oxf) / Year: 2021 Journal: Microscopy (Oxf) / Year: 2021Title: Cryo-EM performance testing of hardware and data acquisition strategies. Authors: Radostin Danev / Haruaki Yanagisawa / Masahide Kikkawa /  Abstract: The increasing popularity and adoption rate of cryo-electron microscopy (cryo-EM) is evidenced by a growing number of new microscope installations around the world. The quality and reliability of the ...The increasing popularity and adoption rate of cryo-electron microscopy (cryo-EM) is evidenced by a growing number of new microscope installations around the world. The quality and reliability of the instruments improved dramatically in recent years, but site-specific issues or unnoticed problems during installation could undermine productivity. Newcomers to the field may also have limited experience and/or low confidence in the capabilities of the equipment or their own skills. Therefore, it is recommended to perform an initial test of the complete cryo-EM workflow with an 'easy' test sample, such as apoferritin, before starting work with real and challenging samples. Analogous test experiments are also recommended for the quantification of new data acquisition approaches or imaging hardware. Here, we present the results from our initial tests of a recently installed Krios G4 electron microscope equipped with two latest generation direct electron detector cameras-Gatan K3 and Falcon 4. Three beam-image shift-based data acquisition strategies were also tested. We detail the methodology and discuss the critical parameters and steps for performance testing. The two cameras performed equally, and the single- and multi-shot per-hole acquisition schemes produced comparable results. We also evaluated the effects of environmental factors and optical flaws on data quality. Our results reaffirmed the exceptional performance of the software aberration correction in Relion in dealing with severe coma aberration. We hope that this work will help cryo-EM teams in their testing and troubleshooting of hardware and data collection approaches. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30686.map.gz emd_30686.map.gz | 446.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30686-v30.xml emd-30686-v30.xml emd-30686.xml emd-30686.xml | 17.5 KB 17.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30686_fsc.xml emd_30686_fsc.xml | 17.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_30686.png emd_30686.png | 151.4 KB | ||
| Masks |  emd_30686_msk_1.map emd_30686_msk_1.map | 476.8 MB |  Mask map Mask map | |
| Others |  emd_30686_half_map_1.map.gz emd_30686_half_map_1.map.gz emd_30686_half_map_2.map.gz emd_30686_half_map_2.map.gz | 375.9 MB 375.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30686 http://ftp.pdbj.org/pub/emdb/structures/EMD-30686 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30686 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30686 | HTTPS FTP |
-Validation report
| Summary document |  emd_30686_validation.pdf.gz emd_30686_validation.pdf.gz | 490.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_30686_full_validation.pdf.gz emd_30686_full_validation.pdf.gz | 490.3 KB | Display | |
| Data in XML |  emd_30686_validation.xml.gz emd_30686_validation.xml.gz | 24.6 KB | Display | |
| Data in CIF |  emd_30686_validation.cif.gz emd_30686_validation.cif.gz | 32.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30686 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30686 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30686 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-30686 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10579 (Title: Krios G4 apoferritin test with K3/(slit out) SerialEM BIS 1x1x4 EMPIAR-10579 (Title: Krios G4 apoferritin test with K3/(slit out) SerialEM BIS 1x1x4Data size: 1.2 TB Data #1: Unaligned multi-frame non-gain-normalized movies in LZW compressed TIFF format [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30686.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30686.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Masked apoferritin map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.599 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
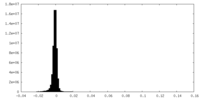
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30686_msk_1.map emd_30686_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
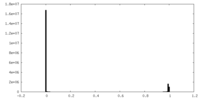
| Density Histograms |
-Half map: Half map 2 corrected for Ewald sphere curvature
| File | emd_30686_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 corrected for Ewald sphere curvature | ||||||||||||
| Projections & Slices |
| ||||||||||||
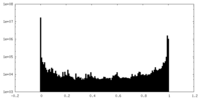
| Density Histograms |
-Half map: Half map 1 corrected for Ewald sphere curvature
| File | emd_30686_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 corrected for Ewald sphere curvature | ||||||||||||
| Projections & Slices |
| ||||||||||||
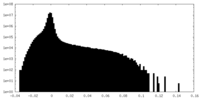
| Density Histograms |
- Sample components
Sample components
-Entire : apoferritin
| Entire | Name: apoferritin |
|---|---|
| Components |
|
-Supramolecule #1: apoferritin
| Supramolecule | Name: apoferritin / type: complex / ID: 1 / Parent: 0 / Details: mouse heavy chain |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 500 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 12 mg/mL |
|---|---|
| Buffer | pH: 7.5 / Details: HEPES-NaOH |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.03 kPa / Details: Harrick Plasma Cleaner on HIGH |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 77.0 K / Max: 77.0 K |
| Alignment procedure | Coma free - Residual tilt: 0.1 mrad |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Digitization - Sampling interval: 5.0 µm / Number grids imaged: 1 / Number real images: 4184 / Average exposure time: 3.04 sec. / Average electron dose: 68.7 e/Å2 Details: Beam-image shift acquisition with a custom SerialEM script. Used SerialEM beam-tilt compensated image shift. Acquisition pattern: 1x1 holes, 4 images/hole |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Calibrated defocus max: 0.8 µm / Calibrated defocus min: 0.2 µm / Calibrated magnification: 125000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 0.8 µm / Nominal defocus min: 0.2 µm / Nominal magnification: 215000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)