[English] 日本語
 Yorodumi
Yorodumi- EMDB-24165: Cryo-EM structure of the human SSU processome, state post-A1 - HE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24165 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the human SSU processome, state post-A1 - HEATR1 focused map | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Vanden Broeck A / Singh S / Klinge S | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Science / Year: 2021 Journal: Science / Year: 2021Title: Nucleolar maturation of the human small subunit processome. Authors: Sameer Singh / Arnaud Vanden Broeck / Linamarie Miller / Malik Chaker-Margot / Sebastian Klinge /  Abstract: The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution ...The human small subunit processome mediates early maturation of the small ribosomal subunit by coupling RNA folding to subsequent RNA cleavage and processing steps. We report the high-resolution cryo–electron microscopy structures of maturing human small subunit (SSU) processomes at resolutions of 2.7 to 3.9 angstroms. These structures reveal the molecular mechanisms that enable crucial progressions during SSU processome maturation. RNA folding states within these particles are communicated to and coordinated with key enzymes that drive irreversible steps such as targeted exosome-mediated RNA degradation, protein-guided site-specific endonucleolytic RNA cleavage, and tightly controlled RNA unwinding. These conserved mechanisms highlight the SSU processome’s impressive structural plasticity, which endows this 4.5-megadalton nucleolar assembly with the distinctive ability to mature the small ribosomal subunit from within. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24165.map.gz emd_24165.map.gz | 37.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24165-v30.xml emd-24165-v30.xml emd-24165.xml emd-24165.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_24165_fsc.xml emd_24165_fsc.xml | 19.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_24165.png emd_24165.png | 124.6 KB | ||
| Masks |  emd_24165_msk_1.map emd_24165_msk_1.map | 669.9 MB |  Mask map Mask map | |
| Others |  emd_24165_additional_1.map.gz emd_24165_additional_1.map.gz emd_24165_half_map_1.map.gz emd_24165_half_map_1.map.gz emd_24165_half_map_2.map.gz emd_24165_half_map_2.map.gz | 28.3 MB 542 MB 542.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24165 http://ftp.pdbj.org/pub/emdb/structures/EMD-24165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24165 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24165 | HTTPS FTP |
-Validation report
| Summary document |  emd_24165_validation.pdf.gz emd_24165_validation.pdf.gz | 419 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24165_full_validation.pdf.gz emd_24165_full_validation.pdf.gz | 418.5 KB | Display | |
| Data in XML |  emd_24165_validation.xml.gz emd_24165_validation.xml.gz | 27.2 KB | Display | |
| Data in CIF |  emd_24165_validation.cif.gz emd_24165_validation.cif.gz | 36.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24165 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24165 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24165 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24165 | HTTPS FTP |
-Related structure data
| Related structure data |  7mq8C  7mq9C  7mqaC  7mqjC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10781 (Title: Nucleolar maturation of the human small subunit processome EMPIAR-10781 (Title: Nucleolar maturation of the human small subunit processomeData size: 74.6 TB Data #1: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 1 [micrographs - multiframe] Data #2: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 2 [micrographs - multiframe] Data #3: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 3 [micrographs - multiframe] Data #4: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 4 [micrographs - multiframe] Data #5: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 5 [micrographs - multiframe] Data #6: Unaligned multi-frame micrograph movies of human SSU processomes - Dataset 6 [micrographs - multiframe] Data #7: Aligned and averaged micrographs of human SSU processomes - Dataset 1 [micrographs - single frame] Data #8: Aligned and averaged micrographs of human SSU processomes - Dataset 2 [micrographs - single frame] Data #9: Aligned and averaged micrographs of human SSU processomes - Dataset 3 [micrographs - single frame] Data #10: Aligned and averaged micrographs of human SSU processomes - Dataset 4 [micrographs - single frame] Data #11: Aligned and averaged micrographs of human SSU processomes - Dataset 5 [micrographs - single frame] Data #12: Aligned and averaged micrographs of human SSU processomes - Dataset 6 [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_24165.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24165.map.gz / Format: CCP4 / Size: 669.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.08 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_24165_msk_1.map emd_24165_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
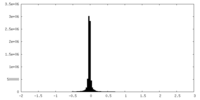
| Density Histograms |
-Additional map: phenix.denmod map
| File | emd_24165_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | phenix.denmod map | ||||||||||||
| Projections & Slices |
| ||||||||||||
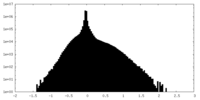
| Density Histograms |
-Half map: #1
| File | emd_24165_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
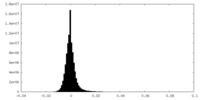
| Density Histograms |
-Half map: #2
| File | emd_24165_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human SSU processome, state post-A1
| Entire | Name: Human SSU processome, state post-A1 |
|---|---|
| Components |
|
-Supramolecule #1: Human SSU processome, state post-A1
| Supramolecule | Name: Human SSU processome, state post-A1 / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 5 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 3.0 nm |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 283 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 84904 / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.7000000000000001 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)