[English] 日本語
 Yorodumi
Yorodumi- EMDB-23313: Negative stain map of monoclonal Fab 23 binding the lateral patch... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23313 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
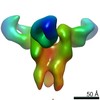
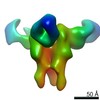
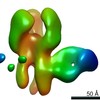
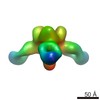
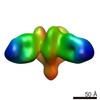
| Title | Negative stain map of monoclonal Fab 23 binding the lateral patch of H1 HA | |||||||||
 Map data Map data | Negative stain map of monoclonal Fab binding the lateral patch of H1 HA | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Han J / Freyn AW / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Influenza hemagglutinin-specific IgA Fc-effector functionality is restricted to stalk epitopes. Authors: Alec W Freyn / Julianna Han / Jenna J Guthmiller / Mark J Bailey / Karlynn Neu / Hannah L Turner / Victoria C Rosado / Veronika Chromikova / Min Huang / Shirin Strohmeier / Sean T H Liu / ...Authors: Alec W Freyn / Julianna Han / Jenna J Guthmiller / Mark J Bailey / Karlynn Neu / Hannah L Turner / Victoria C Rosado / Veronika Chromikova / Min Huang / Shirin Strohmeier / Sean T H Liu / Viviana Simon / Florian Krammer / Andrew B Ward / Peter Palese / Patrick C Wilson / Raffael Nachbagauer /   Abstract: In this study, we utilized a panel of human immunoglobulin (Ig) IgA monoclonal antibodies isolated from the plasmablasts of eight donors after 2014/2015 influenza virus vaccination (Fluarix) to study ...In this study, we utilized a panel of human immunoglobulin (Ig) IgA monoclonal antibodies isolated from the plasmablasts of eight donors after 2014/2015 influenza virus vaccination (Fluarix) to study the binding and functional specificities of this isotype. In this cohort, isolated IgA monoclonal antibodies were primarily elicited against the hemagglutinin protein of the H1N1 component of the vaccine. To compare effector functionalities, an H1-specific subset of antibodies targeting distinct epitopes were expressed as monomeric, dimeric, or secretory IgA, as well as in an IgG1 backbone. When expressed with an IgG Fc domain, all antibodies elicited Fc-effector activity in a primary polymorphonuclear cell-based assay which differs from previous observations that found only stalk-specific antibodies activate the low-affinity FcγRIIIa. However, when expressed with IgA Fc domains, only antibodies targeting the stalk domain showed Fc-effector activity in line with these previous findings. To identify the cause of this discrepancy, we then confirmed that IgG signaling through the high-affinity FcγI receptor was not restricted to stalk epitopes. Since no corresponding high-affinity Fcα receptor exists, the IgA repertoire may therefore be limited to stalk-specific epitopes in the context of Fc receptor signaling. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23313.map.gz emd_23313.map.gz | 11.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23313-v30.xml emd-23313-v30.xml emd-23313.xml emd-23313.xml | 10.8 KB 10.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23313.png emd_23313.png | 62.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23313 http://ftp.pdbj.org/pub/emdb/structures/EMD-23313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23313 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23313 | HTTPS FTP |
-Validation report
| Summary document |  emd_23313_validation.pdf.gz emd_23313_validation.pdf.gz | 296 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23313_full_validation.pdf.gz emd_23313_full_validation.pdf.gz | 295.6 KB | Display | |
| Data in XML |  emd_23313_validation.xml.gz emd_23313_validation.xml.gz | 5.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23313 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23313 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23313 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23313 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23313.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23313.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative stain map of monoclonal Fab binding the lateral patch of H1 HA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.77 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Negative stain map of monoclonal Fab 23 binding the lateral patch...
| Entire | Name: Negative stain map of monoclonal Fab 23 binding the lateral patch of H1 HA |
|---|---|
| Components |
|
-Supramolecule #1: Negative stain map of monoclonal Fab 23 binding the lateral patch...
| Supramolecule | Name: Negative stain map of monoclonal Fab 23 binding the lateral patch of H1 HA type: complex / ID: 1 / Parent: 0 |
|---|
-Supramolecule #2: Monoclonal Fab 23
| Supramolecule | Name: Monoclonal Fab 23 / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F Homo sapiens (human) / Recombinant cell: 293F |
-Supramolecule #3: Hemagglutinin (H1)
| Supramolecule | Name: Hemagglutinin (H1) / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Influenza A virus / Strain: A/California/04/2009(H1N1) Influenza A virus / Strain: A/California/04/2009(H1N1) |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F Homo sapiens (human) / Recombinant cell: 293F |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.02 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Staining | Type: NEGATIVE / Material: uranyl formate |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Average electron dose: 25.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Number images used: 6902 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)






















