[English] 日本語
 Yorodumi
Yorodumi- EMDB-22992: Cryo-EM structure of a thermostable encapsulin from T. maritima -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22992 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a thermostable encapsulin from T. maritima | ||||||||||||
 Map data Map data | Density-modified map made with PHENIX ResolveCryoEM in a 342-pixel box. | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Encapsulin / HK97 fold / flavin-binding / icosahedral / VIRUS LIKE PARTICLE | ||||||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / encapsulin nanocompartment / Hydrolases; Acting on peptide bonds (peptidases) / peptidase activity / iron ion transport / intracellular iron ion homeostasis / proteolysis / Type 1 encapsulin shell protein Function and homology information Function and homology information | ||||||||||||
| Biological species |   Thermotoga maritima MSB8 (bacteria) / Thermotoga maritima MSB8 (bacteria) /   Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria) Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria) | ||||||||||||
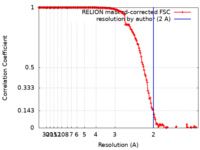
| Method | single particle reconstruction / cryo EM / Resolution: 2.0 Å | ||||||||||||
 Authors Authors | Wiryaman TI / Toor N | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: IUCrJ / Year: 2021 Journal: IUCrJ / Year: 2021Title: Cryo-EM structure of a thermostable bacterial nanocompartment. Authors: Timothy Wiryaman / Navtej Toor /  Abstract: Protein nanocompartments are widespread in bacteria and archaea, but their functions are not yet well understood. Here, the cryo-EM structure of a nanocompartment from the thermophilic bacterium is ...Protein nanocompartments are widespread in bacteria and archaea, but their functions are not yet well understood. Here, the cryo-EM structure of a nanocompartment from the thermophilic bacterium is reported at 2.0 Å resolution. The high resolution of this structure shows that interactions in the E-loop domain may be important for the thermostability of the nanocompartment assembly. Also, the channels at the fivefold axis, threefold axis and dimer interface are assessed for their ability to transport iron. Finally, an unexpected flavin ligand was identified on the exterior of the shell, indicating that this nanocompartment may also play a direct role in iron metabolism. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22992.map.gz emd_22992.map.gz | 141.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22992-v30.xml emd-22992-v30.xml emd-22992.xml emd-22992.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_22992_fsc.xml emd_22992_fsc.xml | 15 KB | Display |  FSC data file FSC data file |
| Images |  emd_22992.png emd_22992.png | 320 KB | ||
| Filedesc metadata |  emd-22992.cif.gz emd-22992.cif.gz | 6.6 KB | ||
| Others |  emd_22992_additional_1.map.gz emd_22992_additional_1.map.gz emd_22992_half_map_1.map.gz emd_22992_half_map_1.map.gz emd_22992_half_map_2.map.gz emd_22992_half_map_2.map.gz | 235.3 MB 236.1 MB 236.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22992 http://ftp.pdbj.org/pub/emdb/structures/EMD-22992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22992 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22992 | HTTPS FTP |
-Validation report
| Summary document |  emd_22992_validation.pdf.gz emd_22992_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22992_full_validation.pdf.gz emd_22992_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_22992_validation.xml.gz emd_22992_validation.xml.gz | 23 KB | Display | |
| Data in CIF |  emd_22992_validation.cif.gz emd_22992_validation.cif.gz | 29.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22992 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22992 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22992 | HTTPS FTP |
-Related structure data
| Related structure data |  7kq5MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10674 (Title: Cryo-EM structure of a thermostable encapsulin from T. maritima EMPIAR-10674 (Title: Cryo-EM structure of a thermostable encapsulin from T. maritimaData size: 1.6 TB Data #1: Unaligned multiframe movies of T. maritima encapsulin [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22992.map.gz / Format: CCP4 / Size: 152.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22992.map.gz / Format: CCP4 / Size: 152.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Density-modified map made with PHENIX ResolveCryoEM in a 342-pixel box. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.7193 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: EM map from RELION Refine3D (note that model...
| File | emd_22992_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EM map from RELION Refine3D (note that model is placed in a 342-pixel box whereas this map is in a 426-pixel box). | ||||||||||||
| Projections & Slices |
| ||||||||||||
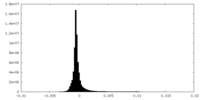
| Density Histograms |
-Half map: Half map 1 from RELION Refine3D
| File | emd_22992_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 from RELION Refine3D | ||||||||||||
| Projections & Slices |
| ||||||||||||
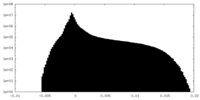
| Density Histograms |
-Half map: Half map 2 from RELION Refine3D
| File | emd_22992_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 from RELION Refine3D | ||||||||||||
| Projections & Slices |
| ||||||||||||
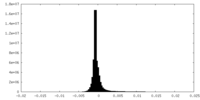
| Density Histograms |
- Sample components
Sample components
-Entire : Encapsulin
| Entire | Name: Encapsulin |
|---|---|
| Components |
|
-Supramolecule #1: Encapsulin
| Supramolecule | Name: Encapsulin / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima MSB8 (bacteria) Thermotoga maritima MSB8 (bacteria) |
| Molecular weight | Theoretical: 1.82868 MDa |
-Macromolecule #1: Maritimacin
| Macromolecule | Name: Maritimacin / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on peptide bonds (peptidases) |
|---|---|
| Source (natural) | Organism:   Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria) Thermotoga maritima (strain ATCC 43589 / MSB8 / DSM 3109 / JCM 10099) (bacteria)Strain: ATCC 43589 / MSB8 / DSM 3109 / JCM 10099 |
| Molecular weight | Theoretical: 30.516787 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEFLKRSFAP LTEKQWQEID NRAREIFKTQ LYGRKFVDVE GPYGWEYAAH PLGEVEVLSD ENEVVKWGLR KSLPLIELRA TFTLDLWEL DNLERGKPNV DLSSLEETVR KVAEFEDEVI FRGCEKSGVK GLLSFEERKI ECGSTPKDLL EAIVRALSIF S KDGIEGPY ...String: MEFLKRSFAP LTEKQWQEID NRAREIFKTQ LYGRKFVDVE GPYGWEYAAH PLGEVEVLSD ENEVVKWGLR KSLPLIELRA TFTLDLWEL DNLERGKPNV DLSSLEETVR KVAEFEDEVI FRGCEKSGVK GLLSFEERKI ECGSTPKDLL EAIVRALSIF S KDGIEGPY TLVINTDRWI NFLKEEAGHY PLEKRVEECL RGGKIITTPR IEDALVVSER GGDFKLILGQ DLSIGYEDRE KD AVRLFIT ETFTFQVVNP EALILLKF UniProtKB: Type 1 encapsulin shell protein |
-Macromolecule #2: FLAVIN MONONUCLEOTIDE
| Macromolecule | Name: FLAVIN MONONUCLEOTIDE / type: ligand / ID: 2 / Number of copies: 1 / Formula: FMN |
|---|---|
| Molecular weight | Theoretical: 456.344 Da |
| Chemical component information |  ChemComp-FMN: |
-Macromolecule #3: water
| Macromolecule | Name: water / type: ligand / ID: 3 / Number of copies: 148 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 12 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: OTHER / Details: Plasma cleaned in Gatan Solarus system | |||||||||
| Vitrification | Cryogen name: PROPANE / Chamber humidity: 90 % / Chamber temperature: 277 K / Instrument: LEICA EM GP Details: 3 ul sample applied to the carbon side of the grid and blotted for 4s before plunging.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 11520 pixel / Digitization - Dimensions - Height: 8184 pixel / Number real images: 2772 / Average exposure time: 2.5 sec. / Average electron dose: 33.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Residue range: 4-269 / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Map was cropped to a 342 box size and density modified with PHENIX ResolveCryoEM |
| Refinement | Protocol: OTHER |
| Output model |  PDB-7kq5: |
 Movie
Movie Controller
Controller














 Z
Z Y
Y X
X