[English] 日本語
 Yorodumi
Yorodumi- EMDB-30130: Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30130 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | |||||||||||||||
 Map data Map data | Post-processed Cryo-EM map generated by RELION | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Nanocompartment / Icosahedral shell / Cargo loaded / VIRUS LIKE PARTICLE | |||||||||||||||
| Function / homology | Type 1 encapsulin shell protein / Encapsulating protein for peroxidase / : / encapsulin nanocompartment / plasma membrane / Type 1 encapsulin shell protein Function and homology information Function and homology information | |||||||||||||||
| Biological species |  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) | |||||||||||||||
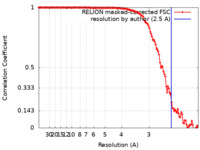
| Method | single particle reconstruction / cryo EM / Resolution: 2.5 Å | |||||||||||||||
 Authors Authors | Tang YT / Mu A / Gong HR / Wang Q / Rao ZH | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2021 Journal: Proc Natl Acad Sci U S A / Year: 2021Title: Cryo-EM structure of DyP-loaded encapsulin. Authors: Yanting Tang / An Mu / Yuying Zhang / Shan Zhou / Weiwei Wang / Yuezheng Lai / Xiaoting Zhou / Fengjiang Liu / Xiuna Yang / Hongri Gong / Quan Wang / Zihe Rao /  Abstract: Encapsulins containing dye-decolorizing peroxidase (DyP)-type peroxidases are ubiquitous among prokaryotes, protecting cells against oxidative stress. However, little is known about how they interact ...Encapsulins containing dye-decolorizing peroxidase (DyP)-type peroxidases are ubiquitous among prokaryotes, protecting cells against oxidative stress. However, little is known about how they interact and function. Here, we have isolated a native cargo-packaging encapsulin from and determined its complete high-resolution structure by cryogenic electron microscopy (cryo-EM). This encapsulin comprises an icosahedral shell and a dodecameric DyP cargo. The dodecameric DyP consists of two hexamers with a twofold axis of symmetry and stretches across the interior of the encapsulin. Our results reveal that the encapsulin shell plays a role in stabilizing the dodecameric DyP. Furthermore, we have proposed a potential mechanism for removing the hydrogen peroxide based on the structural features. Our study also suggests that the DyP is the primary cargo protein of mycobacterial encapsulins and is a potential target for antituberculosis drug discovery. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30130.map.gz emd_30130.map.gz | 167.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30130-v30.xml emd-30130-v30.xml emd-30130.xml emd-30130.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30130_fsc.xml emd_30130_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
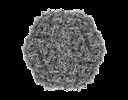
| Images |  emd_30130.png emd_30130.png | 175.3 KB | ||
| Masks |  emd_30130_msk_1.map emd_30130_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-30130.cif.gz emd-30130.cif.gz | 5.7 KB | ||
| Others |  emd_30130_half_map_1.map.gz emd_30130_half_map_1.map.gz emd_30130_half_map_2.map.gz emd_30130_half_map_2.map.gz | 140.7 MB 140.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30130 http://ftp.pdbj.org/pub/emdb/structures/EMD-30130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30130 | HTTPS FTP |
-Related structure data
| Related structure data |  7bojMC  7bokC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30130.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30130.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Post-processed Cryo-EM map generated by RELION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
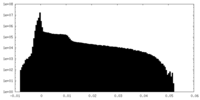
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
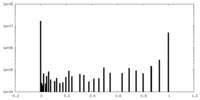
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_30130_msk_1.map emd_30130_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
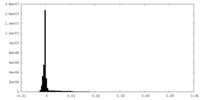
| Density Histograms |
-Half map: First one of the half map after 3D-refinement generated by RELION
| File | emd_30130_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | First one of the half map after 3D-refinement generated by RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
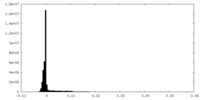
| Density Histograms |
-Half map: Second one of the half map after 3D-refinement generated by RELION
| File | emd_30130_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Second one of the half map after 3D-refinement generated by RELION | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Encapsulin from Mycobacterium smegmatis
| Entire | Name: Encapsulin from Mycobacterium smegmatis |
|---|---|
| Components |
|
-Supramolecule #1: Encapsulin from Mycobacterium smegmatis
| Supramolecule | Name: Encapsulin from Mycobacterium smegmatis / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
-Macromolecule #1: 29 kDa antigen Cfp29
| Macromolecule | Name: 29 kDa antigen Cfp29 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis MC2 155 (bacteria) Mycolicibacterium smegmatis MC2 155 (bacteria) |
| Molecular weight | Theoretical: 28.761162 KDa |
| Sequence | String: MNNLYRDLAP ITESAWAEIE LEATRTFKRH IAGRRVVDVS GPNGPTTASV STGHLLDVSP PGDGVIAHLR DAKPLVRLRV PFTVARRDI DDVERGSQDS DWDPVKDAAK KLAFVEDRAI FEGYAAASIE GIRSSSSNPA LALPDDAREI PDVIAQALSE L RLAGVDGP ...String: MNNLYRDLAP ITESAWAEIE LEATRTFKRH IAGRRVVDVS GPNGPTTASV STGHLLDVSP PGDGVIAHLR DAKPLVRLRV PFTVARRDI DDVERGSQDS DWDPVKDAAK KLAFVEDRAI FEGYAAASIE GIRSSSSNPA LALPDDAREI PDVIAQALSE L RLAGVDGP YSVLLSAETY TKVSETTAHG YPIREHINRL VDGEIIWAPA IDGAFVLSTR GGDFDLQLGT DVSIGYLSHD AE VVHLYME ETMTFLCYTA EASVALTP UniProtKB: Type 1 encapsulin shell protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 10 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-40 / Average exposure time: 6.4 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A / Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
| Output model |  PDB-7boj: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)