+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-22432 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-EM / single particle / protein expression inhibition / RIBOSOME-VIRAL PROTEIN complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationribosomal subunit / negative regulation of RNA splicing / positive regulation of ubiquitin-protein transferase activity / rRNA modification in the nucleus and cytosol / Formation of the ternary complex, and subsequently, the 43S complex / laminin receptor activity / Ribosomal scanning and start codon recognition / Translation initiation complex formation / fibroblast growth factor binding / SARS-CoV-1 modulates host translation machinery ...ribosomal subunit / negative regulation of RNA splicing / positive regulation of ubiquitin-protein transferase activity / rRNA modification in the nucleus and cytosol / Formation of the ternary complex, and subsequently, the 43S complex / laminin receptor activity / Ribosomal scanning and start codon recognition / Translation initiation complex formation / fibroblast growth factor binding / SARS-CoV-1 modulates host translation machinery / Peptide chain elongation / Selenocysteine synthesis / positive regulation of signal transduction by p53 class mediator / Formation of a pool of free 40S subunits / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Eukaryotic Translation Termination / ubiquitin ligase inhibitor activity / Response of EIF2AK4 (GCN2) to amino acid deficiency / SRP-dependent cotranslational protein targeting to membrane / phagocytic cup / Viral mRNA Translation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / 90S preribosome / GTP hydrolysis and joining of the 60S ribosomal subunit / L13a-mediated translational silencing of Ceruloplasmin expression / Major pathway of rRNA processing in the nucleolus and cytosol / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Protein methylation / ribosomal small subunit export from nucleus / laminin binding / rough endoplasmic reticulum / translation regulator activity / gastrulation / Maturation of protein E / MDM2/MDM4 family protein binding / Maturation of protein E / cytosolic ribosome / ER Quality Control Compartment (ERQC) / Myoclonic epilepsy of Lafora / FLT3 signaling by CBL mutants / Prevention of phagosomal-lysosomal fusion / IRAK2 mediated activation of TAK1 complex / Alpha-protein kinase 1 signaling pathway / Glycogen synthesis / IRAK1 recruits IKK complex / IRAK1 recruits IKK complex upon TLR7/8 or 9 stimulation / Membrane binding and targetting of GAG proteins / Endosomal Sorting Complex Required For Transport (ESCRT) / Regulation of TBK1, IKKε (IKBKE)-mediated activation of IRF3, IRF7 / Negative regulation of FLT3 / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / Regulation of TBK1, IKKε-mediated activation of IRF3, IRF7 upon TLR3 ligation / Constitutive Signaling by NOTCH1 HD Domain Mutants / IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation / NOTCH2 Activation and Transmission of Signal to the Nucleus / TICAM1,TRAF6-dependent induction of TAK1 complex / class I DNA-(apurinic or apyrimidinic site) endonuclease activity / TICAM1-dependent activation of IRF3/IRF7 / DNA-(apurinic or apyrimidinic site) lyase / APC/C:Cdc20 mediated degradation of Cyclin B / Regulation of FZD by ubiquitination / Downregulation of ERBB4 signaling / p75NTR recruits signalling complexes / APC-Cdc20 mediated degradation of Nek2A / InlA-mediated entry of Listeria monocytogenes into host cells / TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling / Regulation of innate immune responses to cytosolic DNA / TRAF6-mediated induction of TAK1 complex within TLR4 complex / Regulation of pyruvate metabolism / NF-kB is activated and signals survival / Downregulation of ERBB2:ERBB3 signaling / Pexophagy / NRIF signals cell death from the nucleus / VLDLR internalisation and degradation / Regulation of PTEN localization / Activated NOTCH1 Transmits Signal to the Nucleus / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Regulation of BACH1 activity / MAP3K8 (TPL2)-dependent MAPK1/3 activation / Translesion synthesis by REV1 / TICAM1, RIP1-mediated IKK complex recruitment / Translesion synthesis by POLK / Activation of IRF3, IRF7 mediated by TBK1, IKKε (IKBKE) / Downregulation of TGF-beta receptor signaling / InlB-mediated entry of Listeria monocytogenes into host cell / Josephin domain DUBs / Regulation of activated PAK-2p34 by proteasome mediated degradation / Translesion synthesis by POLI / JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / positive regulation of apoptotic signaling pathway / Gap-filling DNA repair synthesis and ligation in GG-NER / IKK complex recruitment mediated by RIP1 / PINK1-PRKN Mediated Mitophagy / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / TNFR1-induced NF-kappa-B signaling pathway / Autodegradation of Cdh1 by Cdh1:APC/C / APC/C:Cdc20 mediated degradation of Securin / N-glycan trimming in the ER and Calnexin/Calreticulin cycle Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.7 Å | |||||||||
 Authors Authors | Yuan S / Xiong Y | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2020 Journal: Mol Cell / Year: 2020Title: Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. Authors: Shuai Yuan / Lei Peng / Jonathan J Park / Yingxia Hu / Swapnil C Devarkar / Matthew B Dong / Qi Shen / Shenping Wu / Sidi Chen / Ivan B Lomakin / Yong Xiong /  Abstract: The causative virus of the COVID-19 pandemic, SARS-CoV-2, uses its nonstructural protein 1 (Nsp1) to suppress cellular, but not viral, protein synthesis through yet unknown mechanisms. We show here ...The causative virus of the COVID-19 pandemic, SARS-CoV-2, uses its nonstructural protein 1 (Nsp1) to suppress cellular, but not viral, protein synthesis through yet unknown mechanisms. We show here that among all viral proteins, Nsp1 has the largest impact on host viability in the cells of human lung origin. Differential expression analysis of mRNA-seq data revealed that Nsp1 broadly alters the cellular transcriptome. Our cryo-EM structure of the Nsp1-40S ribosome complex shows that Nsp1 inhibits translation by plugging the mRNA entry channel of the 40S. We also determined the structure of the 48S preinitiation complex formed by Nsp1, 40S, and the cricket paralysis virus internal ribosome entry site (IRES) RNA, which shows that it is nonfunctional because of the incorrect position of the mRNA 3' region. Our results elucidate the mechanism of host translation inhibition by SARS-CoV-2 and advance understanding of the impacts from a major pathogenicity factor of SARS-CoV-2. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_22432.map.gz emd_22432.map.gz | 118.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-22432-v30.xml emd-22432-v30.xml emd-22432.xml emd-22432.xml | 48.8 KB 48.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_22432.png emd_22432.png | 141.4 KB | ||
| Filedesc metadata |  emd-22432.cif.gz emd-22432.cif.gz | 11.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-22432 http://ftp.pdbj.org/pub/emdb/structures/EMD-22432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22432 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-22432 | HTTPS FTP |
-Validation report
| Summary document |  emd_22432_validation.pdf.gz emd_22432_validation.pdf.gz | 528.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_22432_full_validation.pdf.gz emd_22432_full_validation.pdf.gz | 528.2 KB | Display | |
| Data in XML |  emd_22432_validation.xml.gz emd_22432_validation.xml.gz | 6.7 KB | Display | |
| Data in CIF |  emd_22432_validation.cif.gz emd_22432_validation.cif.gz | 7.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22432 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22432 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-22432 | HTTPS FTP |
-Related structure data
| Related structure data |  7jqbMC  7jqcC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_22432.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_22432.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.068 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
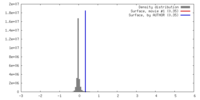
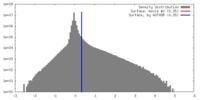
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex
+Supramolecule #1: SARS-CoV-2 Nsp1 and rabbit 40S ribosome complex
+Supramolecule #2: SARS-CoV-2 Nsp1
+Supramolecule #3: rabbit 40S ribosome
+Macromolecule #1: rRNA
+Macromolecule #2: eS25
+Macromolecule #3: 40S ribosomal protein SA
+Macromolecule #4: 40S ribosomal protein S26
+Macromolecule #5: eS1
+Macromolecule #6: uS5
+Macromolecule #7: eS28
+Macromolecule #8: Ribosomal protein S3
+Macromolecule #9: eS30
+Macromolecule #10: uS7
+Macromolecule #11: eS31
+Macromolecule #12: eS6
+Macromolecule #13: RACK1
+Macromolecule #14: eS7
+Macromolecule #15: eS8
+Macromolecule #16: uS4
+Macromolecule #17: S10_plectin domain-containing protein
+Macromolecule #18: eS12
+Macromolecule #19: uS19
+Macromolecule #20: Uncharacterized protein
+Macromolecule #21: eS17
+Macromolecule #22: uS13
+Macromolecule #23: Uncharacterized protein
+Macromolecule #24: uS10
+Macromolecule #25: 40S ribosomal protein S21
+Macromolecule #26: 40S ribosomal protein S24
+Macromolecule #27: Host translation inhibitor nsp1
+Macromolecule #28: 40S ribosomal protein S4
+Macromolecule #29: uS17
+Macromolecule #30: uS15
+Macromolecule #31: Uncharacterized protein
+Macromolecule #32: uS8
+Macromolecule #33: Uncharacterized protein
+Macromolecule #34: eS27
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.7 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 353927 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller




















































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)