10,000 NMR structure milestone reached
10,000 NMR structure milestone reached
The wwPDB is excited to announce that the number of structures available in the PDB archive determined using Nuclear Magnetic Resonance (NMR) spectroscopy has passed the 10,000 mark.
NMR and the PDB
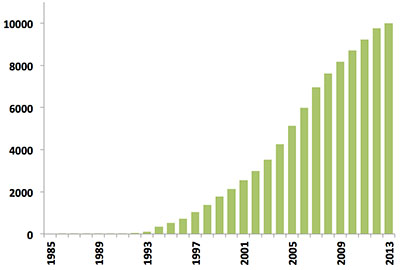
Since the first biomacromolecular NMR structure was archived in 1989, the number of NMR-derived structures in the PDB has grown steadily. Last year alone over 500 new NMR structures were deposited, more than in the first 10 years of NMR depositions combined (Figure 1). Today, NMR-derived structures account for more than 10% of the PDB archive which itself will reach the 100,000 structure mark in 2014.
Figure 1: Yearly growth of released structures in the PDB solved by NMR
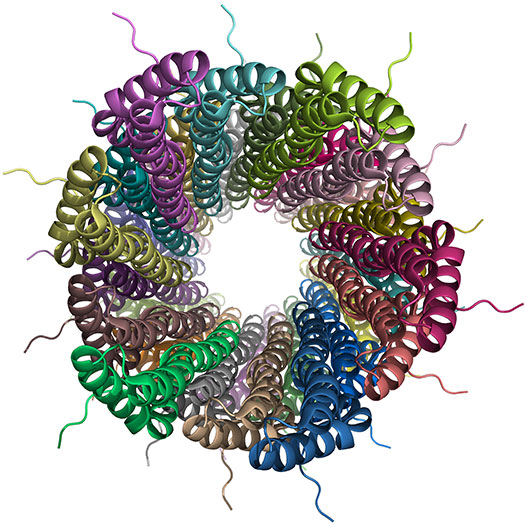
A typical NMR structure in the PDB will consist of an ensemble of models. Nowadays, in addition to the structural ensemble, wwPDB requires the deposition of the assigned chemical shifts as well as the geometric restraints used in the structure determination and refinement. Chemical shifts and restraints are then shared with the BMRB archive , the main repository of experimental NMR data.
NMR structures in the PDB and associated experimental data are presented by the wwPDB partner sites through entry-specific pages. Additionally, a number of dedicated databases, tools and services further help make this wealth of NMR structural information accessible to the scientific community (Table 1).
Future Challenges
As the PDB archive continues to grow, the provision of robust validation tools is becoming even more important. The wwPDB partners are actively driving the development of new standards and software for the validation of structures determined by NMR spectroscopy. A wwPDB NMR Va lidation Task Force (VTF) has been convened to define standard validation criteria, representing community consensus. The VTF has provided its initial recommendations, which will be implemented in a wwPDB validation pipeline and applied to all depositions of NMR structures.
Table 1: Web resources for NMR structures exemplified using PDB entry 2LPZ.
| Description | Resource Link |
|---|---|
| Entry page at PDBe | 2LPZ |
| Entry page at RCSB PDB | 2LPZ |
| Entry page at PDBj | 2LPZ |
| Entry page at BMRB | 18276 2LPZ |
| Vivaldi Visualisation and Validation Information | 2LPZ |
| NMR Restraints Grid | 2LPZ |
| NRG-CING validation information | 2LPZ |
| PSVS validation information | http://psvs-1_4-dev.nesg.org/ |
| ResProx validation information | http://www.resprox.ca/ |
Structure of a bacterial type III secretion needle from Salmonella typhimurium (PDB entry 2LPZ; BMRB entry 18276; Loquet et al., 2012, Nature, 486:276) determined using a hybrid solid-state NMR and electron microscopy (EM) approach.