6PGI
 
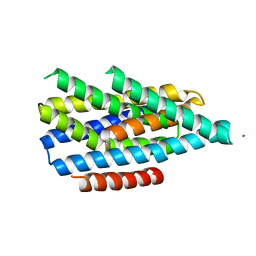 | | Asymmetric functions of a binuclear metal cluster within the transport pathway of the ZIP transition metal transporters | | Descriptor: | BbZIP, CADMIUM ION | | Authors: | Zhang, T, Sui, D, Zhang, C, Logan, T, Hu, J. | | Deposit date: | 2019-06-24 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Asymmetric functions of a binuclear metal center within the transport pathway of a human zinc transporter ZIP4.
Faseb J., 34, 2020
|
|
5Y38
 
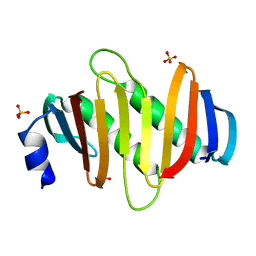 | | Crystal structure of C7orf59-HBXIP complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
4X82
 
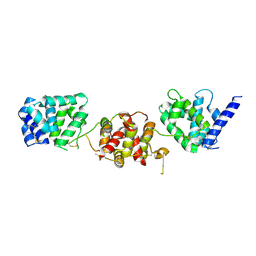 | |
8ZPR
 
 | |
8ZON
 
 | | EcGPR at close conformation. | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-phosphonooxy-pentanoic acid, Gamma-glutamyl phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, T, Leng, Q, Liu, L.J. | | Deposit date: | 2024-05-28 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | EcGPR at close conformation.
To Be Published
|
|
8ZOK
 
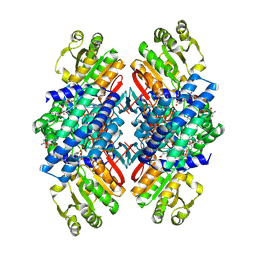 | | EcGPR tetramer with catalytic intermediates | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-phosphonooxy-pentanoic acid, Gamma-glutamyl phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Zhang, T, Leng, Q, Liu, L.J. | | Deposit date: | 2024-05-28 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | EcGPR tetramer with catalytic intermediates
To Be Published
|
|
8ZOO
 
 | | EcGPR at open conformation | | Descriptor: | (2~{S})-2-azanyl-5-oxidanylidene-5-phosphonooxy-pentanoic acid, Gamma-glutamyl phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, T, Leng, Q, Liu, L.J. | | Deposit date: | 2024-05-29 | | Release date: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | EcGPR at open conformation
To Be Published
|
|
4FTX
 
 | | Crystal structure of Ego3 homodimer | | Descriptor: | Protein SLM4, SUCCINIC ACID | | Authors: | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
4FUW
 
 | | Crystal structure of Ego3 mutant | | Descriptor: | Protein SLM4, SULFATE ION | | Authors: | Zhang, T, Peli-Gulli, M.P, Yang, H, De Virgilio, C, Ding, J. | | Deposit date: | 2012-06-28 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ego3 functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the ego complex to activate TORC1.
Structure, 20, 2012
|
|
3DOE
 
 | | Complex of ARL2 and BART, Crystal Form 1 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
3DOF
 
 | | Complex of ARL2 and BART, Crystal Form 2 | | Descriptor: | ADP-ribosylation factor-like protein 2, ADP-ribosylation factor-like protein 2-binding protein, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, T, Li, S, Ding, J. | | Deposit date: | 2008-07-04 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the ARL2-GTP-BART complex reveals a novel recognition and binding mode of small GTPase with effector
Structure, 17, 2009
|
|
5Y39
 
 | | Crystal structure of Ragulator complex (p18 76-145) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
5Y3A
 
 | | Crystal structure of Ragulator complex (p18 49-161) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1.
Nat Commun, 8, 2017
|
|
5TSB
 
 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Cd2+ | | Descriptor: | CADMIUM ION, Membrane protein | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
5TSA
 
 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Zn2+ | | Descriptor: | CADMIUM ION, Membrane protein, ZINC ION | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
8WB3
 
 | |
6JWP
 
 | | crystal structure of EGOC | | Descriptor: | Ego2, GTP-binding protein GTR1, GTP-binding protein GTR2, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2019-04-21 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the EGO-TC-mediated membrane tethering of the TORC1-regulatory Rag GTPases.
Sci Adv, 5, 2019
|
|
9K6K
 
 | | monkeypox thymidine kinase | | Descriptor: | PHOSPHATE ION, THYMIDINE-5'-TRIPHOSPHATE, Thymidine kinase, ... | | Authors: | Zhang, T, Cui, W, Wang, W. | | Deposit date: | 2024-10-22 | | Release date: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.826 Å) | | Cite: | Structural and mechanistic insight into the phosphorylation reaction catalyzed by mpox virus thymidine kinase.
Antiviral Res., 237, 2025
|
|
8J0E
 
 | | GK monomer complexes with catalytic intermediate | | Descriptor: | (2~{R})-5-[[[[(2~{R},3~{S},4~{S},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-2-azanyl-5-oxidanylidene-pentanoic acid, Delta-1-pyrroline-5-carboxylate synthase B, MAGNESIUM ION | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-10 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | GK monomer complexes with catalytic intermediate
To Be Published
|
|
8J0G
 
 | |
8J28
 
 | | GK monomer complexes with G5P and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B, GAMMA-GLUTAMYL PHOSPHATE | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GK monomer complexes with G5P and ADP
To Be Published
|
|
8J27
 
 | | GK monomer complexes with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Delta-1-pyrroline-5-carboxylate synthase B | | Authors: | Zhang, T, Guo, C.J, Liu, J.L. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | GK monomer complexes with ADP
To Be Published
|
|
8ZRI
 
 | |
9AXB
 
 | |
8ZPJ
 
 | |
