1AQZ
 
 | |
1CX5
 
 | | ANTISENSE DNA/RNA HYBRID CONTAINING MODIFIED BACKBONE | | Descriptor: | 5'-D(*CP*GP*CP*GP*TP*T*(MMT)P*TP*GP*CP*GP*C), 5'-R(*GP*CP*GP*CP*AP*AP*AP*AP*CP*GP*CP*G) | | Authors: | Yang, X, Han, X, Cross, C, Sanghvi, Y, Gao, X. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker.
Biochemistry, 38, 1999
|
|
3IR1
 
 | | Crystal Structure of Lipoprotein GNA1946 from Neisseria meningitidis | | Descriptor: | METHIONINE, Outer membrane lipoprotein GNA1946, SULFATE ION | | Authors: | Yang, X, Wu, Z, Wang, X, Shen, Y. | | Deposit date: | 2009-08-21 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of lipoprotein GNA1946 from Neisseria meningitidis
J.Struct.Biol., 168, 2009
|
|
3GXA
 
 | | Crystal structure of GNA1946 | | Descriptor: | METHIONINE, Outer membrane lipoprotein GNA1946, SULFATE ION | | Authors: | Yang, X, Shen, Y. | | Deposit date: | 2009-04-02 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of lipoprotein GNA1946 from Neisseria meningitidis
J.Struct.Biol., 168, 2009
|
|
4ZYL
 
 | |
3LRA
 
 | | Structural Basis for Assembling a Human Tripartite Complex Dlg1-MPP7-Mals3 | | Descriptor: | Disks large homolog 1, MAGUK p55 subfamily member 7, Protein lin-7 homolog C | | Authors: | Yang, X, Xie, X, Shen, Y, Long, J. | | Deposit date: | 2010-02-10 | | Release date: | 2010-11-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for tandem L27 domain-mediated polymerization
Faseb J., 24, 2010
|
|
3DJU
 
 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3DJN
 
 | | Crystal structure of mouse TIS21 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
4YN7
 
 | | Non-oxidized YfiR | | Descriptor: | SULFATE ION, YfiR | | Authors: | Yang, X, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2015-05-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
8J5U
 
 | | Crystal structure of Mycobacterium tuberculosis OppA complexed with an endogenous oligopeptide | | Descriptor: | Endogenous oligopeptide, Uncharacterized protein Rv1280c | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5S
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-catalytic intermediate state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5R
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the resting state | | Descriptor: | IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, Putative peptide transport permease protein Rv1283c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5T
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the catalytic intermediate state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8J5Q
 
 | | Cryo-EM structure of Mycobacterium tuberculosis OppABCD in the pre-translocation state | | Descriptor: | Endogenous oligopeptide, IRON/SULFUR CLUSTER, Putative peptide transport permease protein Rv1282c, ... | | Authors: | Yang, X, Hu, T, Zhang, B, Rao, Z. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | An oligopeptide permease, OppABCD, requires an iron-sulfur cluster domain for functionality.
Nat.Struct.Mol.Biol., 31, 2024
|
|
2BQ0
 
 | | 14-3-3 Protein Beta (Human) | | Descriptor: | 14-3-3 BETA/ALPHA | | Authors: | Yang, X, Elkins, J.M, Fedorov, O, Longman, E.J, Sobott, L, Ball, L.J, Sundstrom, M, Arrowsmith, C, Edwards, A, Doyle, D.A. | | Deposit date: | 2005-04-26 | | Release date: | 2005-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2BR9
 
 | | 14-3-3 Protein Epsilon (Human) Complexed to Peptide | | Descriptor: | 14-3-3 PROTEIN EPSILON, CONSENSUS PEPTIDE FOR 14-3-3 PROTEINS | | Authors: | Yang, X, Elkins, J.M, Soundararajan, M, Fedorov, O, Sundstrom, M, Edwards, A, Arrowsmith, C, Doyle, D.A. | | Deposit date: | 2005-05-03 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Protein-Protein Interactions in the 14-3-3 Protein Family.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
1JBT
 
 | | CRYSTAL STRUCTURE OF RIBOTOXIN RESTRICTOCIN COMPLEXED WITH A 29-MER SARCIN/RICIN DOMAIN RNA ANALOG | | Descriptor: | 29-MER SARCIN/RICIN DOMAIN RNA ANALOG, POTASSIUM ION, RESTRICTOCIN | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBS
 
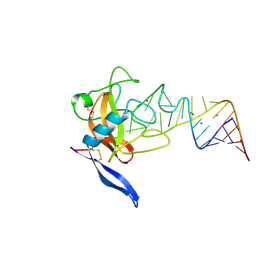 | | Crystal structure of ribotoxin restrictocin and a 29-mer SRD RNA analog | | Descriptor: | 29-mer sarcin/ricin domain RNA analog, POTASSIUM ION, restrictocin | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1JBR
 
 | | Crystal Structure of the Ribotoxin Restrictocin and a 31-mer SRD RNA Inhibitor | | Descriptor: | 31-mer SRD RNA analog, 5'-R(*GP*CP*GP*CP*UP*CP*CP*UP*CP*AP*GP*UP*AP*CP*GP*AP*GP*(A23))-3', 5'-R(*GP*GP*AP*AP*CP*CP*GP*GP*AP*GP*CP*GP*C)-3', ... | | Authors: | Yang, X, Gerczei, T, Glover, L, Correll, C.C. | | Deposit date: | 2001-06-06 | | Release date: | 2001-10-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of restrictocin-inhibitor complexes with implications for RNA recognition and base flipping.
Nat.Struct.Biol., 8, 2001
|
|
1LI4
 
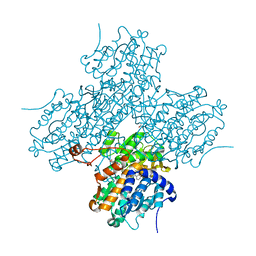 | | Human S-adenosylhomocysteine hydrolase complexed with neplanocin | | Descriptor: | 3-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-CYCLOPENTANE-1,2-DIOL, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, X, Hu, Y, Yin, D.H, Turner, M.A, Wang, M, Borchardt, R.T, Howell, P.L, Kuczera, K, Schowen, R.L. | | Deposit date: | 2002-04-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: Transition-state
stabilization and the avoidance of abortive reactions
Biochemistry, 42, 2003
|
|
8CX4
 
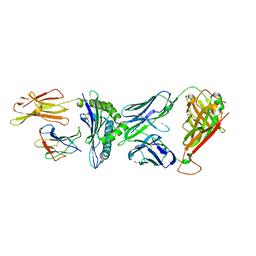 | | TCR-antigen complex AS8.4-YEIH-HLA*B27 | | Descriptor: | AS8.4a, AS8.4b, Beta-2-microglobulin, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-19 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
5B0D
 
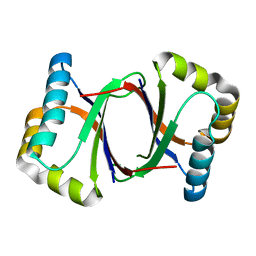 | | Polyketide cyclase OAC from Cannabis sativa, Y27W mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0E
 
 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0F
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0C
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
