7N2O
 
 | | AS4.2-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2Q
 
 | | AS4.3-YEIH-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2S
 
 | | AS3.1-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2R
 
 | | AS4.3-PRPF3-HLA*B27 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2N
 
 | | TCR-antigen complex AS4.2-PRPF3-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
7N2P
 
 | | AS4.3-RNASEH2b-HLA*B27 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AS4.3 T cell receptor alpha chain, AS4.3 T cell receptor beta chain, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2021-05-29 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
1LI4
 
 | | Human S-adenosylhomocysteine hydrolase complexed with neplanocin | | Descriptor: | 3-(6-AMINO-PURIN-9-YL)-5-HYDROXYMETHYL-CYCLOPENTANE-1,2-DIOL, ISOPROPYL ALCOHOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, X, Hu, Y, Yin, D.H, Turner, M.A, Wang, M, Borchardt, R.T, Howell, P.L, Kuczera, K, Schowen, R.L. | | Deposit date: | 2002-04-17 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Catalytic strategy of S-adenosyl-L-homocysteine hydrolase: Transition-state
stabilization and the avoidance of abortive reactions
Biochemistry, 42, 2003
|
|
3DJU
 
 | | Crystal structure of human BTG2 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
3DJN
 
 | | Crystal structure of mouse TIS21 | | Descriptor: | Protein BTG2 | | Authors: | Yang, X, Morita, M, Wang, H, Suzuki, T, Bartlam, M, Yamamoto, T. | | Deposit date: | 2008-06-24 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity
Nucleic Acids Res., 36, 2008
|
|
8CX4
 
 | | TCR-antigen complex AS8.4-YEIH-HLA*B27 | | Descriptor: | AS8.4a, AS8.4b, Beta-2-microglobulin, ... | | Authors: | Yang, X, Jude, K.M, Garcia, K.C. | | Deposit date: | 2022-05-19 | | Release date: | 2022-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Autoimmunity-associated T cell receptors recognize HLA-B*27-bound peptides.
Nature, 612, 2022
|
|
1AQZ
 
 | |
6U9E
 
 | | Structure of PdpA-VgrG Complex, Lidless | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y, Zhou, Z.H, Horwitz, M.A. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
6U9G
 
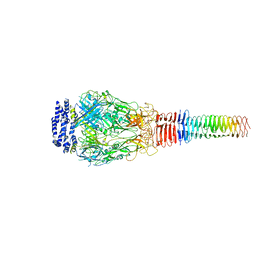 | | Structure of Francisella PdpA-VgrG Complex, half-lidded | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y, Zhou, Z.H, Horwitz, M.A. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
6U9F
 
 | | Structure of Francisella PdpA-VgrG Complex, Lidded | | Descriptor: | PdpA, VgrG | | Authors: | Yang, X, Clemens, D.L, Lee, B.-Y, Cui, Y.X, Horwitz, M.A, Zhou, Z.H. | | Deposit date: | 2019-09-08 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Atomic Structure of the Francisella T6SS Central Spike Reveals a Unique alpha-Helical Lid and a Putative Cargo.
Structure, 27, 2019
|
|
1CX5
 
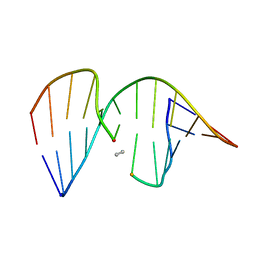 | | ANTISENSE DNA/RNA HYBRID CONTAINING MODIFIED BACKBONE | | Descriptor: | 5'-D(*CP*GP*CP*GP*TP*T*(MMT)P*TP*GP*CP*GP*C), 5'-R(*GP*CP*GP*CP*AP*AP*AP*AP*CP*GP*CP*G) | | Authors: | Yang, X, Han, X, Cross, C, Sanghvi, Y, Gao, X. | | Deposit date: | 1999-08-28 | | Release date: | 1999-09-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an antisense DNA.RNA hybrid duplex containing a 3'-CH(2)N(CH(3))-O-5' or an MMI backbone linker.
Biochemistry, 38, 1999
|
|
4ZYL
 
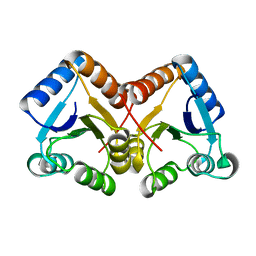 | |
4YN7
 
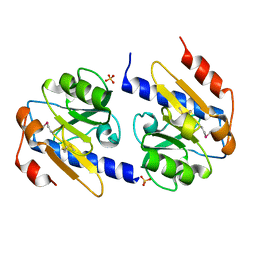 | | Non-oxidized YfiR | | Descriptor: | SULFATE ION, YfiR | | Authors: | Yang, X, Jiang, T. | | Deposit date: | 2015-03-09 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of YfiR from Pseudomonas aeruginosa in two redox states
Biochem.Biophys.Res.Commun., 461, 2015
|
|
9CA1
 
 | | Human TOP3B-TDRD3 core complex in DNA religation state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9C9Y
 
 | | Human TOP3B-TDRD3 core complex in DNA pre-cleavage state | | Descriptor: | DNA (5'-D(P*AP*CP*TP*AP*AP*AP*AP*T)-3'), DNA topoisomerase 3-beta-1, MANGANESE (II) ION, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9CA0
 
 | | Human TOP3B-TDRD3 core complex in DNA post-cleavage state | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*TP*AP*CP*TP*AP*AP*A)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2024-07-17 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9CAH
 
 | | Human TOP3B-TDRD3 complex | | Descriptor: | DNA topoisomerase 3-beta-1, MANGANESE (II) ION, Tudor domain-containing protein 3, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-17 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9CAK
 
 | | Human TOP3B-TDRD3 core complex in DNA rejoining state with ssDNA 5'-GACAGATATT-3 | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*GP*AP*CP*AP*GP*AP*T)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-17 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9CAL
 
 | | Human TOP3B-TDRD3 core complex in post-cleavage state with ssDNA 5'-ACAGATATT-3 | | Descriptor: | DNA (5'-D(*AP*TP*T)-3'), DNA (5'-D(P*GP*AP*CP*AP*GP*AP*T)-3'), DNA topoisomerase 3-beta-1, ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-17 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9CA4
 
 | | Human TOP3B-TDRD3 core complex in RNA rejoining state | | Descriptor: | DNA topoisomerase 3-beta-1, MANGANESE (II) ION, RNA (5'-R(*AP*U)-3'), ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-16 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
9CAG
 
 | | Human TOP3B-TDRD3 core complex in RNA pre-cleavage state | | Descriptor: | DNA topoisomerase 3-beta-1, MANGANESE (II) ION, RNA (5'-R(P*AP*CP*UP*AP*AP*AP*AP*U)-3'), ... | | Authors: | Yang, X, Chen, X, Yang, W, Pommier, Y. | | Deposit date: | 2024-06-17 | | Release date: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural insights into human topoisomerase 3 beta DNA and RNA catalysis and nucleic acid gate dynamics.
Nat Commun, 16, 2025
|
|
