2RTT
 
 | | Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor | | Descriptor: | ChiC | | Authors: | Okumura, A, Uemura, M, Yamada, N, Chikaishi, E, Takai, T, Yoshio, S, Akagi, K, Morita, J, Lee, Y, Yokogawa, D, Suzuki, K, Watanabe, T, Ikegami, T. | | Deposit date: | 2013-08-26 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chitin-binding domain of chitinase Chi18aC from Streptomyces coelicolor
To be Published
|
|
2DCM
 
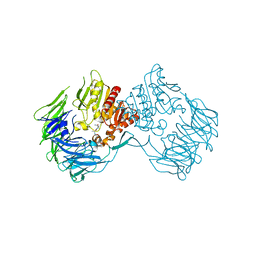 | | The Crystal Structure of S603A Mutated Prolyl Tripeptidyl Aminopeptidase Complexed with Substrate | | Descriptor: | GLYCYLALANYL-N-2-NAPHTHYL-L-PROLINEAMIDE, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-09-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
2D5L
 
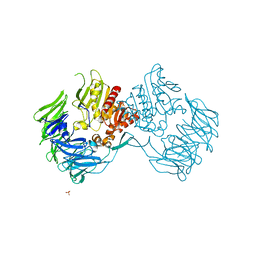 | | Crystal Structure of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis | | Descriptor: | SULFATE ION, dipeptidyl aminopeptidase IV, putative | | Authors: | Nakajima, Y, Ito, K, Xu, Y, Yamada, N, Onohara, Y, Yoshimoto, T. | | Deposit date: | 2005-11-02 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Mechanism of Tripeptidyl Activity of Prolyl Tripeptidyl Aminopeptidase from Porphyromonas gingivalis
J.Mol.Biol., 362, 2006
|
|
5ZFG
 
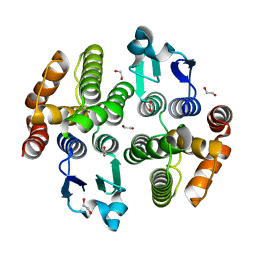 | | Crystal structure of a diazinon-metabolizing glutathione S-transferase in the silkworm, Bombyx mori | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2018-03-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characterisation of a diazinon-metabolising glutathione S-transferase in the silkworm Bombyx mori by X-ray crystallography and genome editing analysis.
Sci Rep, 8, 2018
|
|
3WYW
 
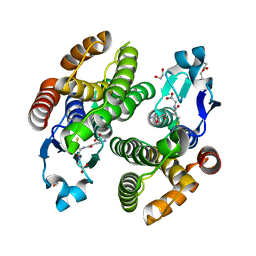 | | Structural characterization of catalytic site of a Nilaparvata lugens delta-class glutathione transferase | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, Glutathione S-transferase | | Authors: | Yamamoto, K, Higashiura, A, Nakagawa, A. | | Deposit date: | 2014-09-09 | | Release date: | 2015-01-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural characterization of the catalytic site of a Nilaparvata lugens delta-class glutathione transferase.
Arch.Biochem.Biophys., 566C, 2014
|
|
3VIR
 
 | | Crystal strcture of Swi5 from fission yeast | | Descriptor: | Mating-type switching protein swi5, octyl beta-D-glucopyranoside | | Authors: | Kuwabara, N, Yamada, N, Hashimoto, H, Sato, M, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
