7RTN
 
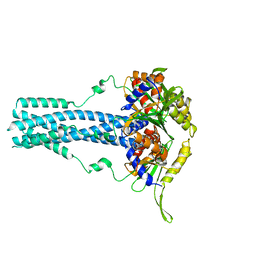 | | Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH | | Descriptor: | Outer capsid protein VP5 | | Authors: | Xia, X, Wu, W.N, Cui, Y.X, Roy, P, Zhou, Z.H. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Bluetongue virus capsid protein VP5 perforates membranes at low endosomal pH during viral entry.
Nat Microbiol, 6, 2021
|
|
7RTO
 
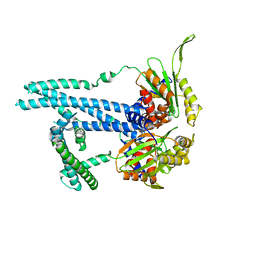 | | Cryo-EM structure of bluetongue virus capsid protein VP5 at low endosomal pH intermediate state 2 | | Descriptor: | Outer capsid protein VP5 | | Authors: | Xia, X, Wu, W.N, Cui, Y.X, Roy, P, Zhou, Z.H. | | Deposit date: | 2021-08-13 | | Release date: | 2021-11-03 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Bluetongue virus capsid protein VP5 perforates membranes at low endosomal pH during viral entry.
Nat Microbiol, 6, 2021
|
|
9E5C
 
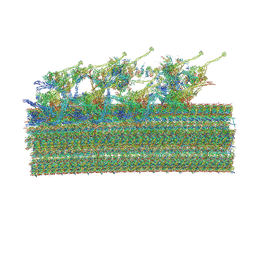 | | Cryo-EM structure of 96 nm repeat of microtubule doublet from T. brucei flagellum | | Descriptor: | 33 kDa inner dynein arm light chain, axonemal, putative, ... | | Authors: | Xia, X, Shimogawa, M.M, Wang, H, Liu, S, Wijono, A, Langousis, G, Kassem, A.M, Wohlschlegel, J.A, Hill, K, Zhou, Z.H. | | Deposit date: | 2024-10-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Trypanosome doublet microtubule structures reveal flagellum assembly and motility mechanisms.
Science, 387, 2025
|
|
9E2G
 
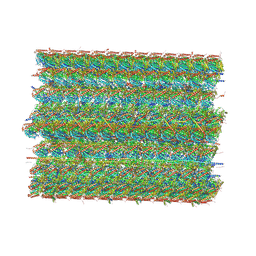 | | Cryo-EM structure of 48 nm repeat of microtubule doublet from T. brucei flagellum | | Descriptor: | CCDC81 HU domain-containing protein, CMF34/CARP4, Calcium-binding protein, ... | | Authors: | Xia, X, Shimogawa, M.M, Wang, H, Liu, S, Wijono, A, Langousis, G, Kassem, A.M, Wohlschlegel, J.A, Hill, K, Zhou, Z.H. | | Deposit date: | 2024-10-22 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Trypanosome doublet microtubule structures reveal flagellum assembly and motility mechanisms.
Science, 387, 2025
|
|
4XKI
 
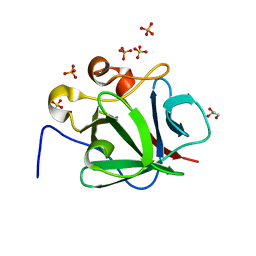 | | Human Fibroblast Growth Factor - 1 (FGF-1) mutant S116R | | Descriptor: | Fibroblast growth factor 1, GLYCEROL, NONAETHYLENE GLYCOL, ... | | Authors: | Xia, X. | | Deposit date: | 2015-01-12 | | Release date: | 2016-01-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biophysical characterizations of Fibroblast Growth Factor - 1 mutant with increased heparin binding affinity
To Be Published
|
|
7TW6
 
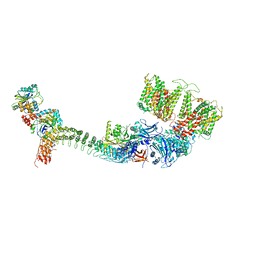 | | Cryo-EM structure of human ankyrin complex (B4P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TVZ
 
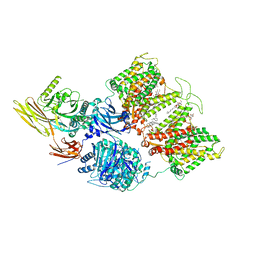 | | Cryo-EM structure of human band 3-protein 4.2 complex in diagonal conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHOLESTEROL, ... | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW3
 
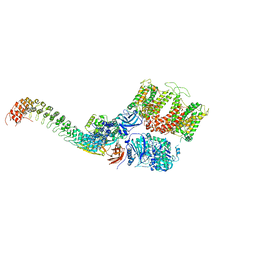 | | Cryo-EM structure of human ankyrin complex (B2P1A1) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW5
 
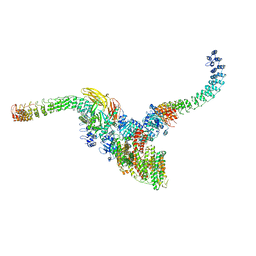 | | Cryo-EM structure of human ankyrin complex (B2P1A2) from red blood cell | | Descriptor: | Ankyrin-1, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TW2
 
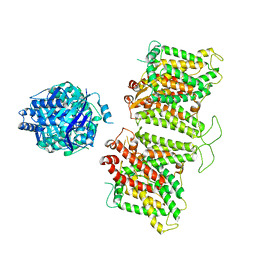 | |
7TW1
 
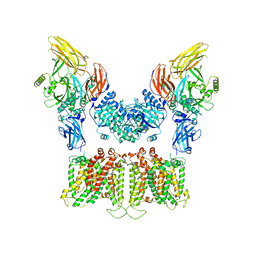 | |
7TW0
 
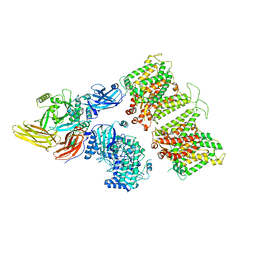 | | Cryo-EM structure of human band 3-protein 4.2 complex in vertical conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, Protein 4.2 | | Authors: | Xia, X, Liu, S.H, Zhou, Z.H. | | Deposit date: | 2022-02-06 | | Release date: | 2022-06-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure, dynamics and assembly of the ankyrin complex on human red blood cell membrane.
Nat.Struct.Mol.Biol., 29, 2022
|
|
4YOL
 
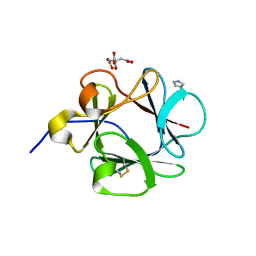 | | Human fibroblast growth factor-1 C16S/A66C/C117A/P134A | | Descriptor: | CITRATE ANION, Fibroblast growth factor 1, IMIDAZOLE | | Authors: | Xia, X, Blaber, M. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Engineering a Cysteine-Free Form of Human Fibroblast Growth Factor-1 for "Second Generation" Therapeutic Application.
J.Pharm.Sci., 105, 2016
|
|
4Q91
 
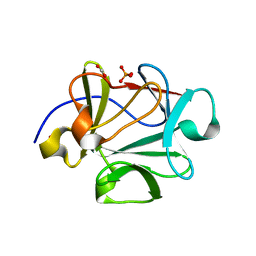 | |
8W1C
 
 | | Cryo-EM structure of BTV pre-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W12
 
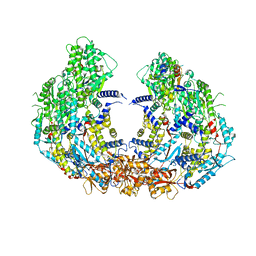 | | Cryo-EM structure of VP3-VP6 heterohexamer | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-14 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1S
 
 | | Cryo-EM structure of BTV pre-core | | Descriptor: | Core protein VP3, RNA-directed RNA polymerase | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1I
 
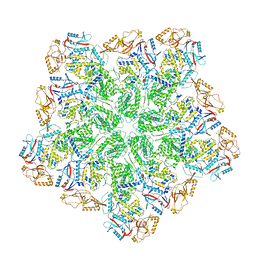 | | Cryo-EM structure of BTV subcore | | Descriptor: | Core protein VP3 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1R
 
 | | Cryo-EM structure of BTV core | | Descriptor: | Core protein VP3, RNA-directed RNA polymerase | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W1O
 
 | | Cryo-EM structure of BTV virion | | Descriptor: | Core protein VP3, Outer capsid protein VP2, RNA-1, ... | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-16 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
8W19
 
 | | Cryo-EM structure of BTV star-subcore | | Descriptor: | Core protein VP3, VP6 | | Authors: | Xia, X, Sung, P.Y, Martynowycz, M.W, Gonen, T, Roy, P, Zhou, Z.H. | | Deposit date: | 2024-02-15 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | RNA genome packaging and capsid assembly of bluetongue virus visualized in host cells.
Cell, 187, 2024
|
|
5K83
 
 | | Crystal Structure of a Primate APOBEC3G N-Domain, in Complex with ssDNA | | Descriptor: | Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G,Apolipoprotein B mRNA editing enzyme, catalytic peptide-like 3G, ... | | Authors: | Xiao, X, Li, S.-X, Yang, H, Chen, X.S. | | Deposit date: | 2016-05-27 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of APOBEC3G N-domain alone and its complex with DNA.
Nat Commun, 7, 2016
|
|
9NEZ
 
 | | Structure of the Pyrococcus furiosus SHI complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Xiao, X, Li, H. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus.
Structure, 2025
|
|
9NF0
 
 | | Structure of the NADPH-bound Pyrococcus furiosus SHI complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Xiao, X, Li, H. | | Deposit date: | 2025-02-20 | | Release date: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural insights into the biotechnologically relevant reversible NADPH-oxidizing NiFe-hydrogenase from P.furiosus.
Structure, 2025
|
|
8D6Y
 
 | |
