6O0H
 
 | |
5TRC
 
 | |
5CS4
 
 | |
5CS0
 
 | |
5CSK
 
 | |
5CSL
 
 | |
5CSA
 
 | |
4DZD
 
 | |
3JUI
 
 | | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit | | Descriptor: | GLYCEROL, Translation initiation factor eIF-2B subunit epsilon | | Authors: | Wei, J, Xu, H, Zhang, C, Wang, M, Gao, F, Gong, W. | | Deposit date: | 2009-09-15 | | Release date: | 2009-10-06 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the C-terminal Domain of Human Translation Initiation Factor eIF2B epsilon Subunit
To be published
|
|
2K6S
 
 | |
7VZN
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZQ
 
 | | The structure of GdmN V24Y/G157A/R158A/G188R mutant in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, CHLORIDE ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZU
 
 | | The structure of GdmN Y82F mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GdmN, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYP
 
 | | The structure of GdmN complex with the natural tetrahedral intermediate, carbamoylated derivative, and AMP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, FE (III) ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZY
 
 | | The structure of GdmN complex with AMP and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYJ
 
 | | The structure of GdmN in complex with carbamoyl adenylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VYO
 
 | | The structure of GdmN | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VZZ
 
 | | The structure of GdmN in complex with the natural tetrahedral intermediate, carbamoyl adenylate, and 20-O-methyl-19-chloroproansamitocin | | Descriptor: | (5~{S},6~{E},8~{S},9~{S},12~{R},15~{E})-21-chloranyl-12,20-dimethoxy-6,8,16-trimethyl-5,9-bis(oxidanyl)-2-azabicyclo[16.3.1]docosa-1(21),6,15,18(22),19-pentaene-3,11-dione, 1,2-ETHANEDIOL, 5'-O-[(S)-(carbamoyloxy)(hydroxy)phosphoryl]adenosine, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7VX0
 
 | | The structure of GdmN complex with ATP | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, FE (III) ION, ... | | Authors: | Wei, J, Zheng, J, Zhou, J, Kang, Q, Bai, L. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Endowing homodimeric carbamoyltransferase GdmN with iterative functions through structural characterization and mechanistic studies.
Nat Commun, 13, 2022
|
|
7PAB
 
 | | Varicella zoster Orf24-Orf27 nuclear egress complex | | Descriptor: | Nuclear egress protein 2,Nuclear egress protein 1, SULFATE ION, ZINC ION | | Authors: | Schweininger, J, Muller, Y.A. | | Deposit date: | 2021-07-29 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the varicella-zoster Orf24-Orf27 nuclear egress complex spotlights multiple determinants of herpesvirus subfamily specificity.
J.Biol.Chem., 298, 2022
|
|
7OJU
 
 | | Chaetomium thermophilum Naa50 GNAT-domain in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | CARBOXYMETHYL COENZYME *A, GLYCEROL, HEXAETHYLENE GLYCOL, ... | | Authors: | Weidenhausen, J, Kopp, J, Sinning, I. | | Deposit date: | 2021-05-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Extended N-Terminal Acetyltransferase Naa50 in Filamentous Fungi Adds to Naa50 Diversity.
Int J Mol Sci, 23, 2022
|
|
6Z00
 
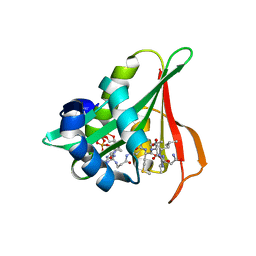 | | Arabidopsis thaliana Naa50 in complex with bisubstrate analogue CoA-Ac-MVNAL | | Descriptor: | Acyl-CoA N-acyltransferases (NAT) superfamily protein, CARBOXYMETHYL COENZYME *A, MET-VAL-ASN-ALA-LEU | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
6YZZ
 
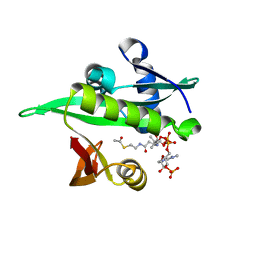 | | Arabidopsis thaliana Naa50 in complex with AcCoA | | Descriptor: | ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Weidenhausen, J, Kopp, J, Lapouge, K, Sinning, I. | | Deposit date: | 2020-05-07 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and functional characterization of the N-terminal acetyltransferase Naa50.
Structure, 29, 2021
|
|
1BQJ
 
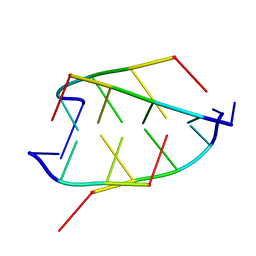 | | CRYSTAL STRUCTURE OF D(ACCCT) | | Descriptor: | DNA (5'-D(*AP*CP*CP*CP*T)-3') | | Authors: | Weil, J, Min, T, Yang, C, Wang, S, Sutherland, C, Sinha, N, Kang, C.H. | | Deposit date: | 1998-08-17 | | Release date: | 1999-03-18 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Stabilization of the i-motif by intramolecular adenine-adenine-thymine base triple in the structure of d(ACCCT).
Acta Crystallogr.,Sect.D, 55, 1999
|
|
6TQD
 
 | | D00-D0 domain of Intimin | | Descriptor: | BROMIDE ION, CHLORIDE ION, Intimin, ... | | Authors: | Weikum, J, Leo, J.C, Morth, J.P. | | Deposit date: | 2019-12-16 | | Release date: | 2021-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The extracellular juncture domains in the intimin passenger adopt a constitutively extended conformation inducing restraints to its sphere of action.
Sci Rep, 10, 2020
|
|
