6T40
 
 | | Bovine enterovirus F3 in complex with a Cysteinylglycine dipeptide | | Descriptor: | CHLORIDE ION, CYSTEINE, GLYCEROL, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
6T3F
 
 | | Crystal structure Nipah virus fusion glycoprotein in complex with a neutralising Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab66 heavy chain, ... | | Authors: | Avanzato, V.A, Pryce, R, Walter, T.S, Bowden, T.A. | | Deposit date: | 2019-10-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for antibody-mediated neutralization of Nipah virus reveals a site of vulnerability at the fusion glycoprotein apex.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6T48
 
 | | Bovine enterovirus F3 in complex with glutathione and a Cysteinylglycine dipeptide | | Descriptor: | CHLORIDE ION, CYSTEINE, GLUTATHIONE, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-12 | | Release date: | 2020-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
2VD9
 
 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) with bound L-Ala-P | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, ALANINE RACEMASE, CHLORIDE ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2V8O
 
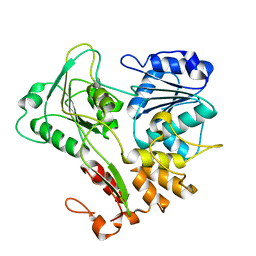 | | Structure of the Murray Valley encephalitis virus RNA helicase to 1. 9A resolution | | Descriptor: | FLAVIVIRIN PROTEASE NS3 | | Authors: | Mancini, E.J, Assenberg, R, Verma, A, Walter, T.S, Tuma, R, Grimes, J.M, Owens, R.J, Stuart, D.I. | | Deposit date: | 2007-08-09 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Murray Valley Encephalitis Virus RNA Helicase at 1.9 A Resolution.
Protein Sci., 16, 2007
|
|
2UVD
 
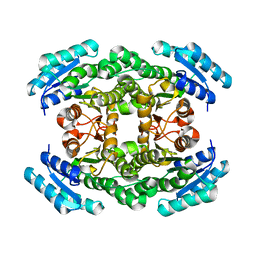 | | The crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase from Bacillus anthracis (BA3989) | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER-PROTEIN) REDUCTASE | | Authors: | Zaccai, N.R, Carter, L.G, Berrow, N.S, Sainsbury, S, Nettleship, J.E, Walter, T.S, Harlos, K, Owens, R.J, Wilson, K.S, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-03-09 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of a 3-Oxoacyl-(Acylcarrier Protein) Reductase (Ba3989) from Bacillus Anthracis at 2.4-A Resolution.
Proteins: Struct., Funct., Bioinf., 70, 2008
|
|
2VD8
 
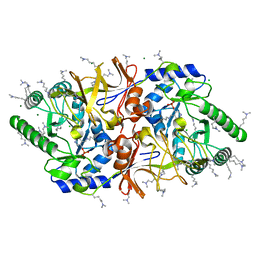 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) | | Descriptor: | ALANINE RACEMASE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
5MQW
 
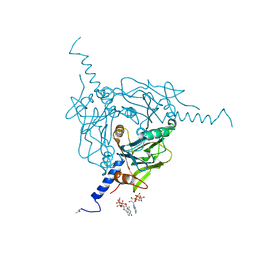 | | High-speed fixed-target serial virus crystallography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
5OSN
 
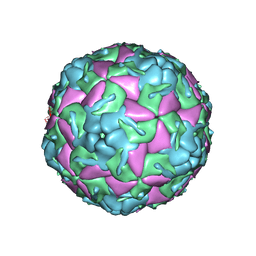 | | Crystal Structure of Bovine Enterovirus 2 determined with Serial Femtosecond X-ray Crystallography | | Descriptor: | Capsid protein, GLUTAMIC ACID, POTASSIUM ION, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Kotecha, A, Kelly, J, Rowlands, D.J, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
6QGI
 
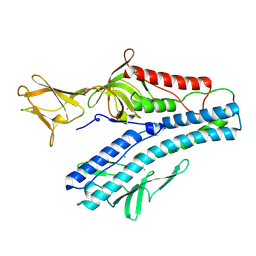 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
6QGL
 
 | | Crystal structure of VP5 from Haloarchaeal pleomorphic virus 6 | | Descriptor: | BROMIDE ION, VP5 | | Authors: | El Omari, K, Walter, T.S, Harlos, K, Grimes, J.M, Stuart, D.I, Roine, E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The structure of a prokaryotic viral envelope protein expands the landscape of membrane fusion proteins.
Nat Commun, 10, 2019
|
|
6QT9
 
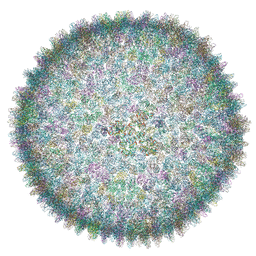 | | Cryo-EM structure of SH1 full particle. | | Descriptor: | ORF 24, ORF 25, ORF 31, ... | | Authors: | De Colibus, L, Roine, E, Walter, T.S, Ilca, S.L, Wang, X, Wang, N, Roseman, A.M, Bamford, D, Huiskonen, J.T, Stuart, D.I. | | Deposit date: | 2019-02-22 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Assembly of complex viruses exemplified by a halophilic euryarchaeal virus.
Nat Commun, 10, 2019
|
|
6T4C
 
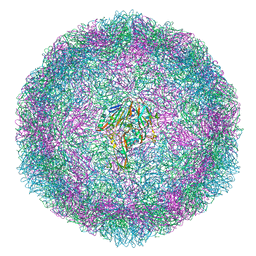 | | Bovine enterovirus F3 in complex with glutathione | | Descriptor: | GLUTATHIONE, GLYCEROL, POTASSIUM ION, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Walter, T.S, Fry, E.E, Stuart, D.I. | | Deposit date: | 2019-10-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glutathione facilitates enterovirus assembly by binding at a druggable pocket.
Commun Biol, 3, 2020
|
|
4FJV
 
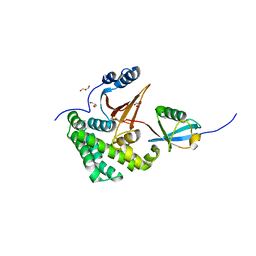 | | Crystal Structure of Human Otubain2 and Ubiquitin Complex | | Descriptor: | ETHANAMINE, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Altun, M, Walter, T.S, Kramer, H.B, Iphofer, A, David, Y, Komsany, A, Ternette, N, Nicholson, B, Navon, A, Stuart, D.I, Ren, J, Kessler, B.M. | | Deposit date: | 2012-06-12 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The human otubain2-ubiquitin structure provides insights into the cleavage specificity of poly-ubiquitin-linkages.
Plos One, 10, 2015
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDX
 
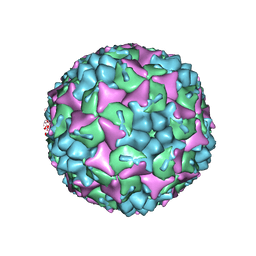 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP12 | | Descriptor: | 1-(5-((3'-METHYL-[1,1'-BIPHENYL]-4-YL)OXY)PENTYL)-3-(, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-07 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDW
 
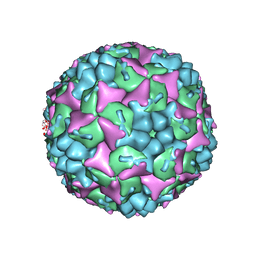 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP4 | | Descriptor: | 1-[(3S)-5-(4-iodanylphenoxy)-3-methyl-pentyl]-3-pyridin-4-yl-imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDQ
 
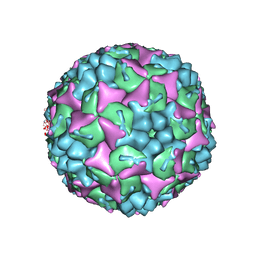 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CEW
 
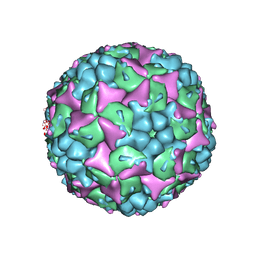 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor ALD | | Descriptor: | 4-[3-[(3s)-5-[4-[(e)-ethoxyiminomethyl]phenoxy]-3-methyl-pentyl]-2-oxidanylidene-imidazolidin-1-yl]pyridine-2-carboxamide, VP1, VP2, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4CDU
 
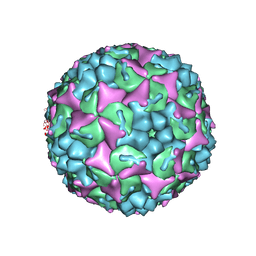 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, CHLORIDE ION, SODIUM ION, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-06 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
3G3Z
 
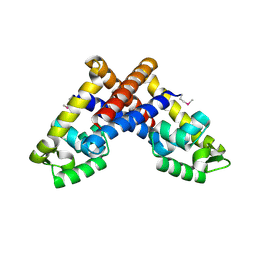 | | The structure of NMB1585, a MarR family regulator from Neisseria meningitidis | | Descriptor: | Transcriptional regulator, MarR family | | Authors: | Nichols, C.E, Sainsbury, S, Ren, J, Walter, T.S, Verma, A, Stammers, D.K, Saunders, N.J, Owens, R.J, Oxford Protein Production Facility (OPPF) | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of NMB1585, a MarR-family regulator from Neisseria meningitidis
Acta Crystallogr.,Sect.F, 65, 2009
|
|
5WTH
 
 | | Cryo-EM structure for Hepatitis A virus complexed with FAB | | Descriptor: | FAB Heavy Chain, FAB Light Chain, Polyprotein, ... | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTF
 
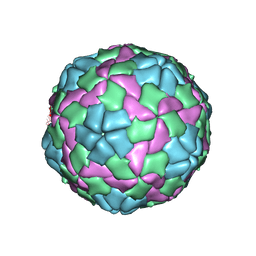 | | Cryo-EM structure for Hepatitis A virus empty particle | | Descriptor: | VP0, VP1, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTE
 
 | | Cryo-EM structure for Hepatitis A virus full particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WTG
 
 | | Crystal structure of the Fab fragment of anti-HAV antibody R10 | | Descriptor: | FAB Heavy chain, FAB Light chain | | Authors: | Wang, X, Zhu, L, Dang, M, Hu, Z, Gao, Q, Yuan, S, Sun, Y, Zhang, B, Ren, J, Walter, T.S, Wang, J, Fry, E.E, Stuart, D.I, Rao, Z. | | Deposit date: | 2016-12-11 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | Potent neutralization of hepatitis A virus reveals a receptor mimic mechanism and the receptor recognition site
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
