1FZT
 
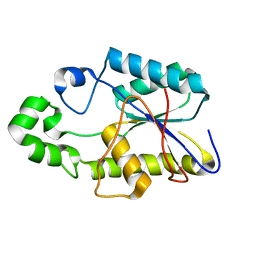 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
1DV9
 
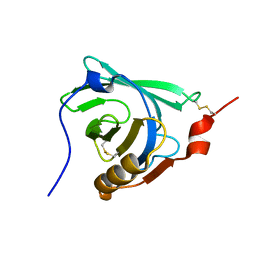 | | STRUCTURAL CHANGES ACCOMPANYING PH-INDUCED DISSOCIATION OF THE B-LACTOGLOBULIN DIMER | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Uhrinova, S, Smith, M.H, Jameson, G.B, Uhrin, D, Sawyer, L, Barlow, P.N. | | Deposit date: | 2000-01-20 | | Release date: | 2000-02-09 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural changes accompanying pH-induced dissociation of the beta-lactoglobulin dimer.
Biochemistry, 39, 2000
|
|
1NWV
 
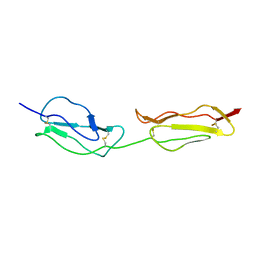 | | SOLUTION STRUCTURE OF A FUNCTIONALLY ACTIVE COMPONENT OF DECAY ACCELERATING FACTOR | | Descriptor: | Complement decay-accelerating factor | | Authors: | Uhrinova, S, Lin, F, Ball, G, Bromek, K, Uhrin, D, Medof, M.E, Barlow, P.N. | | Deposit date: | 2003-02-07 | | Release date: | 2003-04-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a functionally active fragment of decay-accelerating factor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1Z1M
 
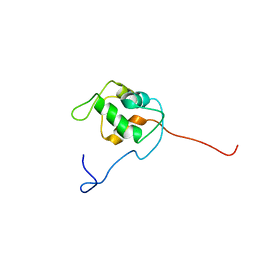 | | NMR structure of unliganded MDM2 | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Uhrinova, S, Uhrin, D, Powers, H, Watt, K, Zheleva, D, Fischer, P, McInnes, C, Barlow, P.N. | | Deposit date: | 2005-03-04 | | Release date: | 2005-06-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of Free MDM2 N-terminal Domain Reveals Conformational Adjustments that Accompany p53-binding
J.Mol.Biol., 350, 2005
|
|
1PPQ
 
 | | NMR structure of 16th module of Immune Adherence Receptor, Cr1 (Cd35) | | Descriptor: | Complement receptor type 1 | | Authors: | O'Leary, J.M, Bromek, K, Black, G.M, Uhrinova, S, Schmitz, C, Krych, M, Atkinson, J.P, Uhrin, D, Barlow, P.N. | | Deposit date: | 2003-06-17 | | Release date: | 2004-05-04 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics of complement control protein (CCP) modules reveals mobility in binding surfaces.
Protein Sci., 13, 2004
|
|
