2NRD
 
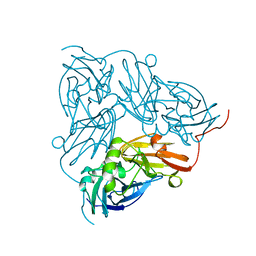 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
2GQ2
 
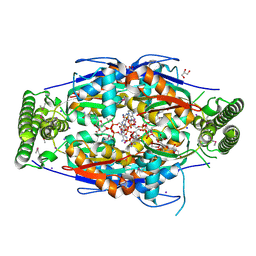 | | Mycobacterium tuberculosis ThyX-NADP complex | | Descriptor: | GLYCEROL, IODIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sampathkumar, P, Turley, S, Sibley, C.H, Hol, W.G. | | Deposit date: | 2006-04-19 | | Release date: | 2006-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | NADP+ expels both the co-factor and a substrate analog from the Mycobacterium tuberculosis ThyX active site: opportunities for anti-bacterial drug design.
J.Mol.Biol., 360, 2006
|
|
1AS6
 
 | |
1AS8
 
 | |
1AQ8
 
 | |
1AS7
 
 | |
2AFN
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE AND A COPPER SITE MUTANT, M150E, THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
6BQM
 
 | | Secreted serine protease VesC from Vibrio cholerae | | Descriptor: | serine protease VesC | | Authors: | Park, Y.J, Korotkov, K.V, Delarosa, J.R, Turley, S, DiMaio, F, Hol, W.G.J. | | Deposit date: | 2017-11-28 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Suppressor Mutations in Type II Secretion Mutants of Vibrio cholerae: Inactivation of the VesC Protease.
Msphere, 5, 2020
|
|
1RL4
 
 | | Plasmodium falciparum peptide deformylase complex with inhibitor | | Descriptor: | (2R)-2-{[FORMYL(HYDROXY)AMINO]METHYL}HEXANOIC ACID, 2-{N'-[2-(5-AMINO-1-PHENYLCARBAMOYL-PENTYLCARBAMOYL)-HEXYL]-HYDRAZINOMETHYL}-HEXANOIC ACID(5-AMINO-1-PHENYLCARBAMOYL-PENTYL)-AMIDE, COBALT (II) ION, ... | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G.J. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase.
Protein Sci., 13, 2004
|
|
1RQC
 
 | | Crystals of peptide deformylase from Plasmodium falciparum with ten subunits per asymmetric unit reveal critical characteristics of the active site for drug design | | Descriptor: | COBALT (II) ION, formylmethionine deformylase | | Authors: | Robien, M.A, Nguyen, K.T, Kumar, A, Hirsh, I, Turley, S, Pei, D, Hol, W.G. | | Deposit date: | 2003-12-04 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An improved crystal form of Plasmodium falciparum peptide deformylase
Protein Sci., 13, 2004
|
|
1JYM
 
 | | Crystals of Peptide Deformylase from Plasmodium falciparum with Ten Subunits per Asymmetric Unit Reveal Critical Characteristics of the Active Site for Drug Design | | Descriptor: | COBALT (II) ION, Peptide Deformylase | | Authors: | Kumar, A, Nguyen, K.T, Srivathsan, S, Ornstein, B, Turley, S, Hirsh, I, Pei, D, Hol, W.G.J. | | Deposit date: | 2001-09-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystals of peptide deformylase from Plasmodium falciparum reveal critical characteristics of the active site for drug design.
Structure, 10, 2002
|
|
1NTD
 
 | | STRUCTURE OF ALCALIGENES FAECALIS NITRITE REDUCTASE MUTANT M150E THAT CONTAINS ZINC | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Murphy, M.E.P, Adman, E.T, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Alcaligenes faecalis nitrite reductase and a copper site mutant, M150E, that contains zinc.
Biochemistry, 34, 1995
|
|
1NIE
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIC
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE, SULFATE ION | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NID
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIA
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIF
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
1NIB
 
 | | THE STRUCTURE OF CU-NITRITE REDUCTASE FROM ACHROMOBACTER CYCLOCLASTES AT FIVE PH VALUES, WITH NITRITE BOUND AND WITH TYPE II CU DEPLETED | | Descriptor: | COPPER (II) ION, NITRITE REDUCTASE | | Authors: | Adman, E.T, Godden, J.W, Turley, S. | | Deposit date: | 1995-07-03 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2- bound and with type II copper depleted.
J.Biol.Chem., 270, 1995
|
|
2QAC
 
 | | The closed MTIP-MyosinA-tail complex from the malaria parasite invasion machinery | | Descriptor: | Myosin A tail domain interacting protein MTIP, Myosin-A | | Authors: | Bosch, J, Turley, S, Roach, C.M, Daly, T.M, Bergman, L.W, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-06-14 | | Release date: | 2007-06-26 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Closed MTIP-Myosin A-Tail Complex from the Malaria Parasite Invasion Machinery.
J.Mol.Biol., 372, 2007
|
|
4GGN
 
 | | Malaria invasion machinery protein complex | | Descriptor: | Myosin A tail domain interacting protein MTIP, Myosin-A | | Authors: | Khamrui, S, Turley, S, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
4GFT
 
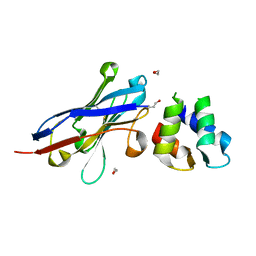 | | Malaria invasion machinery protein-Nanobody complex | | Descriptor: | 1,2-ETHANEDIOL, Myosin A tail domain interacting protein, Nanobody | | Authors: | Khamrui, S, Turley, S, Pardon, E, Steyaert, J, Verlinde, C, Fan, E, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-03 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
4KSS
 
 | | Crystal Structure of Vibrio cholerae ATPase GspsE Hexamer | | Descriptor: | Type II secretion system protein E, hemolysin-coregulated protein | | Authors: | Hol, W.G, Turley, S, Lu, C.Y, Park, Y.J. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (7.58 Å) | | Cite: | Hexamers of the Type II Secretion ATPase GspE from Vibrio cholerae with Increased ATPase Activity.
Structure, 21, 2013
|
|
4KSR
 
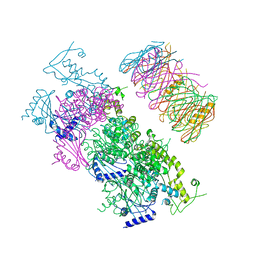 | | Crystal Structure of the Vibrio cholerae ATPase GspE Hexamer | | Descriptor: | Type II secretion system protein E, Hemolysin-coregulated protein | | Authors: | Hol, W.G, Turley, S, Lu, C.Y, Park, Y.J, Marionni, S.T, Lee, K, Patrick, M, Sandkvist, M, Bush, M, Shah, R. | | Deposit date: | 2013-05-17 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Hexamers of the Type II Secretion ATPase GspE from Vibrio cholerae with Increased ATPase Activity.
Structure, 21, 2013
|
|
1QS0
 
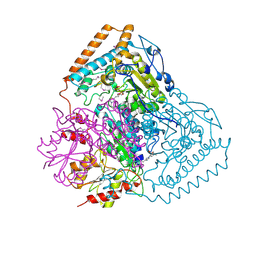 | | Crystal Structure of Pseudomonas Putida 2-oxoisovalerate Dehydrogenase (Branched-Chain Alpha-Keto Acid Dehydrogenase, E1B) | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-OXOISOVALERATE DEHYDROGENASE ALPHA-SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA-SUBUNIT, ... | | Authors: | Aevarsson, A, Seger, K, Turley, S, Sokatch, J.R, Hol, W.G.J. | | Deposit date: | 1999-06-24 | | Release date: | 1999-08-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of 2-oxoisovalerate and dehydrogenase and the architecture of 2-oxo acid dehydrogenase multienzyme complexes.
Nat.Struct.Biol., 6, 1999
|
|
1HXI
 
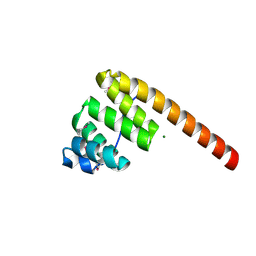 | | AN UNEXPECTED EXTENDED CONFORMATION FOR THE THIRD TPR MOTIF OF THE PEROXIN PEX5 FROM TRYPANOSOMA BRUCEI | | Descriptor: | MAGNESIUM ION, PEROXISOME TARGETING SIGNAL 1 RECEPTOR PEX5 | | Authors: | Kumar, A, Roach, C, Hirsh, I.S, Turley, S, deWalque, S, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2001-01-15 | | Release date: | 2001-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unexpected extended conformation for the third TPR motif of the peroxin PEX5 from Trypanosoma brucei.
J.Mol.Biol., 307, 2001
|
|
