1I18
 
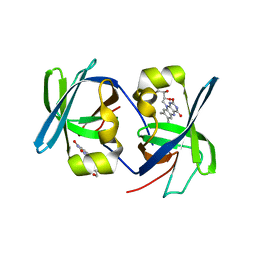 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-31 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1HZE
 
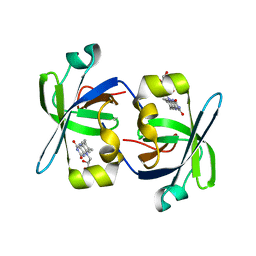 | | SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF RIBOFLAVIN SYNTHASE FROM E. COLI | | Descriptor: | RIBOFLAVIN, RIBOFLAVIN SYNTHASE ALPHA CHAIN | | Authors: | Truffault, V, Coles, M, Diercks, T, Abelmann, K, Eberhardt, S, Luettgen, H, Bacher, A, Kessler, H. | | Deposit date: | 2001-01-24 | | Release date: | 2001-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of riboflavin synthase.
J.Mol.Biol., 309, 2001
|
|
1YFB
 
 | |
2RM4
 
 | |
1YSF
 
 | |
2BZE
 
 | | NMR Structure of human RTF1 PLUS3 domain. | | Descriptor: | KIAA0252 PROTEIN | | Authors: | Truffault, V, Diercks, T, Ab, E, De Jong, R.N, Daniels, M.A, Kaptein, R, Folkers, G.E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-08-16 | | Release date: | 2007-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA Binding of the Human Rtf1 Plus3 Domain.
Structure, 16, 2008
|
|
2LYD
 
 | |
2GLW
 
 | | The solution structure of PHS018 from pyrococcus horikoshii | | Descriptor: | 92aa long hypothetical protein | | Authors: | Coles, M, Hulko, M, Truffault, V, Martin, J, Lupas, A.N. | | Deposit date: | 2006-04-05 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Common evolutionary origin of swapped-hairpin and double-psi beta barrels
Structure, 14, 2006
|
|
6QH2
 
 | |
2P3M
 
 | | Solution structure of Mj0056 | | Descriptor: | Riboflavin Kinase MJ0056 | | Authors: | Coles, M, Truffault, V, Djuranovic, S, Martin, J, Lupas, A.N. | | Deposit date: | 2007-03-09 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A CTP-Dependent Archaeal Riboflavin Kinase Forms a Bridge in the Evolution of Cradle-Loop Barrels.
Structure, 15, 2007
|
|
6QF8
 
 | |
6QFP
 
 | |
1W6V
 
 | | Solution structure of the DUSP domain of hUSP15 | | Descriptor: | UBIQUITIN CARBOXYL-TERMINAL HYDROLASE 15 | | Authors: | De Jong, R.D, Ab, E, Diercks, T, Truffault, V, Daniels, M, Kaptein, R, Folkers, G.E. | | Deposit date: | 2004-08-24 | | Release date: | 2006-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Human Ubiquitin-Specific Protease 15 Dusp Domain.
J.Biol.Chem., 281, 2006
|
|
2M3X
 
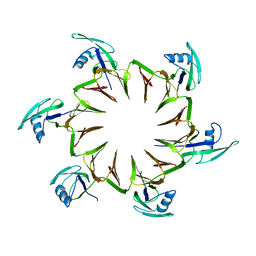 | |
4A53
 
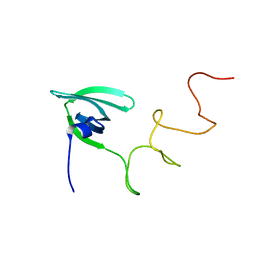 | | Structural basis of the Dcp1:Dcp2 mRNA decapping complex activation by Edc3 and Scd6 | | Descriptor: | EDC3 | | Authors: | Fromm, S.A, Truffault, V, Kamenz, J, Braun, J.E, Hoffmann, N.A, Izaurralde, E, Sprangers, R. | | Deposit date: | 2011-10-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Edc3- and Scd6-Mediated Activation of the Dcp1:Dcp2 Mrna Decapping Complex.
Embo J., 31, 2011
|
|
4A54
 
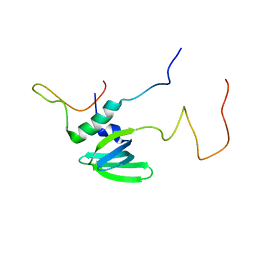 | | Structural basis of the Dcp1:Dcp2 mRNA decapping complex activation by Edc3 and Scd6 | | Descriptor: | EDC3, MRNA DECAPPING COMPLEX SUBUNIT 2 | | Authors: | Fromm, S.A, Truffault, V, Kamenz, J, Braun, J.E, Hoffmann, N.A, Izaurralde, E, Sprangers, R. | | Deposit date: | 2011-10-24 | | Release date: | 2012-02-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Structural Basis of Edc3- and Scd6-Mediated Activation of the Dcp1:Dcp2 Mrna Decapping Complex.
Embo J., 31, 2011
|
|
2BW2
 
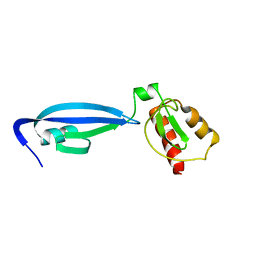 | | BofC from Bacillus subtilis | | Descriptor: | BYPASS OF FORESPORE C | | Authors: | Patterson, H.M, Brannigan, J.A, Cutting, S.M, Wilson, K.S, Wilkinson, A.J, Ab, E, Diercks, T, Folkers, G.E, de Jong, R.N, Truffault, V, Kaptein, R. | | Deposit date: | 2005-07-08 | | Release date: | 2005-09-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of bypass of forespore C, an intercompartmental signaling factor during sporulation in Bacillus.
J. Biol. Chem., 280, 2005
|
|
2LLE
 
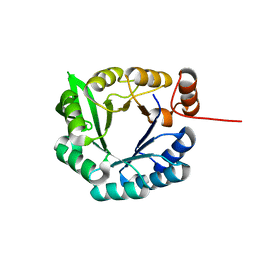 | | Computational design of an eight-stranded (beta/alpha)-barrel from fragments of different folds | | Descriptor: | Chemotaxis protein CheY, Imidazole glycerol phosphate synthase subunit HisF chimera | | Authors: | Coles, M, Truffault, V, Eisenbeis, S, Proffitt, W, Meiler, J, Hocker, B. | | Deposit date: | 2011-11-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Potential of fragment recombination for rational design of proteins.
J.Am.Chem.Soc., 134, 2012
|
|
2LDY
 
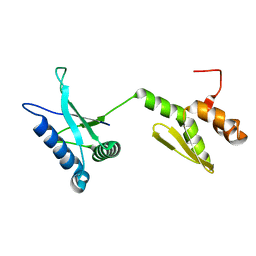 | |
2JV2
 
 | |
5A66
 
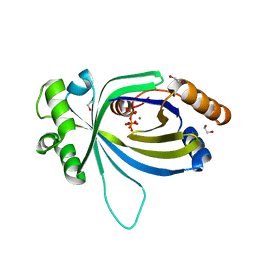 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and manganese ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE (II) ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A61
 
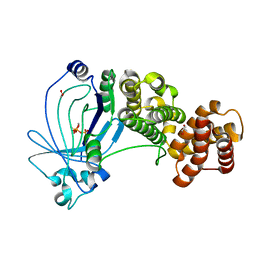 | | Crystal structure of full-length E. coli ygiF in complex with tripolyphosphate and two manganese ions. | | Descriptor: | 1,2-ETHANEDIOL, INORGANIC TRIPHOSPHATASE, MANGANESE (II) ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A5Y
 
 | | Crystal structure of AtTTM3 in complex with tripolyphosphate and magnesium ion (form A) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TRIPHOSPHATE, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A64
 
 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine triphosphate. | | Descriptor: | 1,2-ETHANEDIOL, THIAMINE TRIPHOSPHATASE, TRIETHYLENE GLYCOL, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
5A65
 
 | | Crystal structure of mouse thiamine triphosphatase in complex with thiamine diphosphate, orthophosphate and magnesium ions. | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Martinez, J, Truffault, V, Hothorn, M. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Determinants for Substrate Binding and Catalysis in Triphosphate Tunnel Metalloenzymes.
J.Biol.Chem., 290, 2015
|
|
